A proton-coupled dynamic conformational switch in the HIV-1 dimerization initiation site kissing complex
- PMID: 14734802
- PMCID: PMC337028
- DOI: 10.1073/pnas.0307966100
A proton-coupled dynamic conformational switch in the HIV-1 dimerization initiation site kissing complex
Abstract
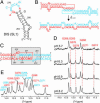
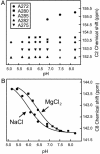
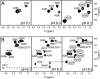
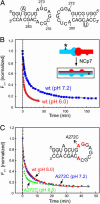
In HIV type 1 (HIV-1), the dimerization initiation site (DIS) is the sequence primarily responsible for initiating the noncovalent linkage of two homologous strands of genomic RNA during viral assembly. The DIS loop contains an autocomplementary hexanucleotide sequence and forms a symmetric homodimer through a loop-loop kissing interaction. In a structural rearrangement catalyzed by the HIV-1 nucleocapsid protein (NCp7) and suggested to be associated with maturation of the budded viral particle, the DIS converts from a metastable kissing dimer to an extended duplex. Here, we demonstrate that the DIS kissing dimer displays localized conformational dynamics that result from the specific protonation of the N1 base nitrogen of the DIS loop residue A272 at near-physiological pH. The rate of NCp7-catalyzed maturation of the DIS kissing dimer is also shown to directly correlate with the observed proton-coupled conformational dynamics, where NCp7 is found to convert the dynamic A272 protonated state with a faster rate. Taken together, these results reveal a previously undescribed role for base protonation in modulating local RNA structure and demonstrate a mechanism for promoting the chaperone-mediated structural rearrangement of a kinetically trapped RNA conformational state.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

