A genomewide oscillation in transcription gates DNA replication and cell cycle
- PMID: 14734811
- PMCID: PMC337030
- DOI: 10.1073/pnas.0306490101
A genomewide oscillation in transcription gates DNA replication and cell cycle
Abstract
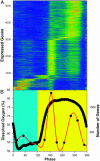
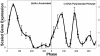
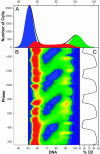
Microarray analysis from a yeast continuous synchrony culture system shows a genomewide oscillation in transcription. Maximums in transcript levels occur at three nearly equally spaced intervals in this approximately 40-min cycle of respiration and reduction. Two temporal clusters (4,679 of 5,329) are maximally expressed during the reductive phase of the cycle, whereas a third cluster (650) is maximally expressed during the respiratory phase. Transcription is organized functionally into redox-state superclusters with genes known to be important in respiration or reduction being synthesized in opposite phases of the cycle. The transcriptional cycle gates synchronous bursts in DNA replication in a constant fraction of the population at 40-min intervals. Restriction of DNA synthesis to the reductive phase of the cycle may be an evolutionarily important mechanism for reducing oxidative damage to DNA during replication.
Figures



Comment in
-
An ultradian clock shapes genome expression in yeast.Proc Natl Acad Sci U S A. 2004 Feb 3;101(5):1118-9. doi: 10.1073/pnas.0400052101. Epub 2004 Jan 26. Proc Natl Acad Sci U S A. 2004. PMID: 14745022 Free PMC article. No abstract available.
References
-
- Klevecz, R. R. & Ruddle, F. H. (1968) Science 159, 634–636. - PubMed
-
- Kauffman, S. A. & Wille, J. J. (1975) J. Theor. Biol. 55, 47–93. - PubMed
-
- Mackey, M. C. & Glass, L. (1977) Science 197, 287–289. - PubMed
-
- Waddington, C. D. (1957) The Strategy of the Gene (Allen and Unwin, London)
-
- Eigen, M. & Schuster, P. (1977) Naturwissenschaften 64, 541–565. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

