RNase H2 of Saccharomyces cerevisiae is a complex of three proteins
- PMID: 14734815
- PMCID: PMC373335
- DOI: 10.1093/nar/gkh209
RNase H2 of Saccharomyces cerevisiae is a complex of three proteins
Erratum in
- Nucleic Acids Res. 2004 Feb 24;32(4):1616
Abstract
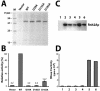
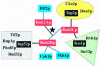
The composition of RNase H2 has been a long-standing problem. Whereas bacterial and archaeal RNases H2 are active as single polypeptides, the Saccharomyces cerevisiae homolog, Rnh2Ap, when expressed in Escherichia coli, fails to produce an active RNase H2. By affinity chromatography purification and identification of polypeptides associated with a tagged S.cerevisiae Rnh2Ap, we obtained a complex of three proteins [Rnh2Ap (Rnh201p), Ydr279p (Rnh202p) and Ylr154p (Rnh203p)] that together are necessary and sufficient for RNase H2 activity [correction]. Deletion of the gene encoding any one of the proteins or mutations in the catalytic site in Rnh2A led to loss of RNase H2 activity. Even when S.cerevisiae RNase H2 is catalytically compromised, it still exhibits a preference for cleavage of the phosphodiester bond on the 5' side of a ribonucleotide-deoxyribonucleotide sequence in substrates mimicking RNA-primed Okazaki fragments or a single ribonucleotide embedded in a duplex DNA. Interestingly, Ydr279p and Ylr154p have homologous proteins only in closely related species. The multisubunit nature of S.cerevisiae RNase H2 may be important both for structural purposes and to provide a means of interacting with other proteins involved in DNA replication/repair and transcription.
Figures



References
-
- Crouch R.J. and Cerritelli,S.M. (1998) RNases H of S.cerevisiae, S.pombe, C.fasciculata, and N.crassa. In Crouch,R.J. and Toulmé,J.J. (eds), Ribonucleases H. INSERM, Paris, pp. 79–100.
-
- Ohtani N., Haruki,M., Morikawa,M., Crouch,R.J., Itaya,M. and Kanaya,S. (1999) Identification of the genes encoding Mn2+-dependent RNase I–III and Mg2+-dependent RNase HIII from Bacillus subtilis: classification of RNases H into three families. Biochemistry, 38, 605–618. - PubMed
-
- Chapados B.R., Chai,Q., Hosfield,D.J., Qiu,J.Z., Shen,B.H. and Tainer,J.A. (2001) Structural biochemistry of a type 2 RNase H: RNA primer recognition and removal during DNA replication. J. Mol. Biol., 307, 541–556. - PubMed
-
- Ohtani N., Haruki,M., Morikawa,M. and Kanaya,S. (1999) Molecular diversities of RNases H. J. Biosci. Bioeng., 88, 12–19. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

