A hydration study of (1-->4) and (1-->6) linked alpha-glucans by comparative 10 ns molecular dynamics simulations and 500-MHz NMR
- PMID: 14735575
- PMCID: PMC4201036
- DOI: 10.1002/jcc.10405
A hydration study of (1-->4) and (1-->6) linked alpha-glucans by comparative 10 ns molecular dynamics simulations and 500-MHz NMR
Abstract
The hydration behavior of two model disaccharides, methyl-alpha-D-maltoside (1) and methyl-alpha-D-isomaltoside (2), has been investigated by a comparative 10 ns molecular dynamics study. The detailed hydration of the two disaccharides was described using three force fields especially developed for modeling of carbohydrates in explicit solvent. To validate the theoretical results the two compounds were synthesized and subjected to 500 MHz NMR spectroscopy, including pulsed field gradient diffusion measurements (1: 4.0. 10(-6) cm(2). s(-1); 2: 4.2. 10(-6) cm(2). s(-1)). In short, the older CHARMM-based force field exhibited a more structured carbohydrate-water interaction leading to better agreement with the diffusional properties of the two compounds, whereas especially the alpha-(1-->6) linkage and the primary hydroxyl groups were inaccurately modeled. In contrast, the new generation of the CHARMM-based force field (CSFF) and the most recent version of the AMBER-based force field (GLYCAM-2000a) exhibited less structured carbohydrate-water interactions with the result that the diffusional properties of the two disaccharides were underestimated, whereas the simulations of the alpha-(1-->6) linkage and the primary hydroxyl groups were significantly improved and in excellent agreement with homo- and heteronuclear coupling constants. The difference between the two classes of force field (more structured and less structured carbohydrate-water interaction) was underlined by calculation of the isotropic hydration as calculated by radial pair distributions. At one extreme, the radial O em leader O pair distribution function yielded a peak density of 2.3 times the bulk density in the first hydration shell when using the older CHARMM force field, whereas the maximum density observed in the GLYCAM force field was calculated to be 1.0, at the other extreme.
Copyright 2004 Wiley Periodicals, Inc. J Comput Chem 25: 573-586, 2004
Figures

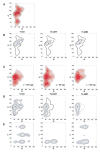


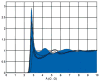
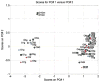


References
-
- Brady JW. J Am Chem Soc. 1989;111:5155.
-
- Engelsen SB, Hervé du Penhoat C, Pérez S. J Phys Chem. 1995;99:13334.
-
- Engelsen SB, Monteiro C, Hervé du Penhoat C, Pérez S. Biophys Chem. 2001;93:103. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

