Protein interaction networks by proteome peptide scanning
- PMID: 14737190
- PMCID: PMC314469
- DOI: 10.1371/journal.pbio.0020014
Protein interaction networks by proteome peptide scanning
Abstract
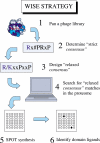
A substantial proportion of protein interactions relies on small domains binding to short peptides in the partner proteins. Many of these interactions are relatively low affinity and transient, and they impact on signal transduction. However, neither the number of potential interactions mediated by each domain nor the degree of promiscuity at a whole proteome level has been investigated. We have used a combination of phage display and SPOT synthesis to discover all the peptides in the yeast proteome that have the potential to bind to eight SH3 domains. We first identified the peptides that match a relaxed consensus, as deduced from peptides selected by phage display experiments. Next, we synthesized all the matching peptides at high density on a cellulose membrane, and we probed them directly with the SH3 domains. The domains that we have studied were grouped by this approach into five classes with partially overlapping specificity. Within the classes, however, the domains display a high promiscuity and bind to a large number of common targets with comparable affinity. We estimate that the yeast proteome contains as few as six peptides that bind to the Abp1 SH3 domain with a dissociation constant lower than 100 microM, while it contains as many as 50-80 peptides with corresponding affinity for the SH3 domain of Yfr024c. All the targets of the Abp1 SH3 domain, identified by this approach, bind to the native protein in vivo, as shown by coimmunoprecipitation experiments. Finally, we demonstrate that this strategy can be extended to the analysis of the entire human proteome. We have developed an approach, named WISE (whole interactome scanning experiment), that permits rapid and reliable identification of the partners of any peptide recognition module by peptide scanning of a proteome. Since the SPOT synthesis approach is semiquantitative and provides an approximation of the dissociation constants of the several thousands of interactions that are simultaneously analyzed in an array format, the likelihood of each interaction occurring in any given physiological settings can be evaluated. WISE can be easily extended to a variety of protein interaction domains, including those binding to modified peptides, thereby offering a powerful proteomic tool to help completing a full description of the cell interactome.
Conflict of interest statement
The authors have declared that no conflicts of interest exist.
Figures






References
-
- Aasland R, Abrams C, Ampe C, Ball LJ, Bedford MT, et al. Normalization of nomenclature for peptide motifs as ligands of modular protein domains. FEBS Lett. 2002;513:141–144. - PubMed
-
- Cestra G, Castagnoli L, Dente L, Minenkova O, Petrelli A, et al. The SH3 domains of endophilin and amphiphysin bind to the proline-rich region of synaptojanin 1 at distinct sites that display an unconventional binding specificity. J Biol Chem. 1999;274:32001–32007. - PubMed
-
- de Heuvel E, Bell AW, Ramjaun AR, Wong K, Sossin WS, et al. Identification of the major synaptojanin-binding proteins in brain. J Biol Chem. 1997;272:8710–8716. - PubMed
-
- Fazi B, Cope MJ, Douangamath A, Ferracuti S, Schirwitz K, et al. Unusual binding properties of the SH3 domain of the yeast actin-binding protein Abp1: Structural and functional analysis. J Biol Chem. 2002;277:5290–5298. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

