Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds
- PMID: 14737219
- PMCID: PMC314300
- DOI: 10.1371/journal.pbio.0020007
Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds
Abstract
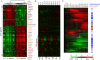
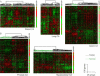
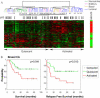
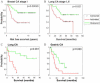
Cancer invasion and metastasis have been likened to wound healing gone awry. Despite parallels in cellular behavior between cancer progression and wound healing, the molecular relationships between these two processes and their prognostic implications are unclear. In this study, based on gene expression profiles of fibroblasts from ten anatomic sites, we identify a stereotyped gene expression program in response to serum exposure that appears to reflect the multifaceted role of fibroblasts in wound healing. The genes comprising this fibroblast common serum response are coordinately regulated in many human tumors, allowing us to identify tumors with gene expression signatures suggestive of active wounds. Genes induced in the fibroblast serum-response program are expressed in tumors by the tumor cells themselves, by tumor-associated fibroblasts, or both. The molecular features that define this wound-like phenotype are evident at an early clinical stage, persist during treatment, and predict increased risk of metastasis and death in breast, lung, and gastric carcinomas. Thus, the transcriptional signature of the response of fibroblasts to serum provides a possible link between cancer progression and wound healing, as well as a powerful predictor of the clinical course in several common carcinomas.
Conflict of interest statement
The authors have declared that no conflicts of interest exist.
Figures
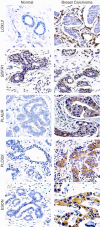
References
-
- Akiri G, Sabo E, Dafni H, Vadasz Z, Kartvelishvily Y, et al. Lysyl oxidase-related protein-1 promotes tumor fibrosis and tumor progression in vivo . Cancer Res. 2003;63:1657–1666. - PubMed
-
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

