Disabling the folding catalyst is the last critical step in alpha-lytic protease folding
- PMID: 14739318
- PMCID: PMC2286698
- DOI: 10.1110/ps.03389704
Disabling the folding catalyst is the last critical step in alpha-lytic protease folding
Abstract
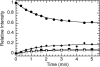
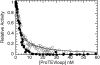
Alpha-Lytic protease (alphaLP) is an extracellular bacterial pro-protease marked by extraordinary conformational rigidity and a highly cooperative barrier to unfolding. Although these properties successfully limit its proteolytic destruction, thereby extending the functional lifetime of the protease, they come at the expense of foldability (t(1/2) = 1800 yr) and thermodynamic stability (native alphaLP is less stable than the unfolded species). Efficient folding has required the coevolution of a large N-terminal pro region (Pro) that rapidly catalyzes alphaLP folding (t(1/2) = 23 sec) and shifts the thermodynamic equilibrium in favor of folded protease through tight native-state binding. Release of active alphaLP from this stabilizing, but strongly inhibitory, complex requires the proteolytic destruction of Pro. alphaLP is capable of initiating Pro degradation via cleavage of a flexible loop within the Pro C-terminal domain. This single cleavage event abolishes Pro catalysis while maintaining strong native-state binding. Thus, the loop acts as an Achilles' heel by which the Pro foldase machinery can be safely dismantled, preventing Pro-catalyzed unfolding, without compromising alphaLP native-state stability. Once the loop is cleaved, Pro is rapidly degraded, releasing active alphaLP.
Figures



References
-
- Baker, D., Sohl, J.L., and Agard, D.A. 1992. A protein-folding reaction under kinetic control. Nature 356 263–265. - PubMed
-
- Baker, D., Shiau, A.K., and Agard, D.A. 1993. The role of pro regions in protein folding. Curr. Opin. Cell Biol. 5 966–970. - PubMed
-
- Brayer, G.D., Delbaere, L.T.J., and James, M.N.G. 1979. Molecular structure of the α-lytic protease from Myxobacter 495 at 2 Å resolution. J. Mol. Biol. 131 743–775. - PubMed
-
- Cunningham, E.L. and Agard, D.A. 2003. Interdependent folding of the N- and C-terminal domains defines the cooperative folding of α-lytic protease. Biochemistry 42 13212–13219. - PubMed
-
- Cunningham, E.L., Mau, T., Truhlar, S.M.E., and Agard, D.A. 2002. The pro region N-terminal domain provides specific interactions required for catalysis of α-lytic protease folding. Biochemistry 41 8860–8867. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

