Both rare and common polymorphisms contribute functional variation at CHGA, a regulator of catecholamine physiology
- PMID: 14740315
- PMCID: PMC1181918
- DOI: 10.1086/381399
Both rare and common polymorphisms contribute functional variation at CHGA, a regulator of catecholamine physiology
Abstract
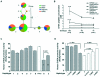
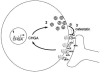
The chromogranin/secretogranin proteins are costored and coreleased with catecholamines from secretory vesicles in chromaffin cells and noradrenergic neurons. Chromogranin A (CHGA) regulates catecholamine storage and release through intracellular (vesiculogenic) and extracellular (catecholamine release-inhibitory) mechanisms. CHGA is a candidate gene for autonomic dysfunction syndromes, including intermediate phenotypes that contribute to human hypertension. Here, we show a surprising pattern of CHGA variants that alter the expression and function of this gene, both in vivo and in vitro. Functional variants include both common alleles that quantitatively alter gene expression and rare alleles that qualitatively change the encoded product to alter the signaling potency of CHGA-derived catecholamine release-inhibitory catestatin peptides.
Figures


Similar articles
-
The chromogranin A fragment catestatin: specificity, potency and mechanism to inhibit exocytotic secretion of multiple catecholamine storage vesicle co-transmitters.J Hypertens. 2006 May;24(5):895-904. doi: 10.1097/01.hjh.0000222760.99852.e0. J Hypertens. 2006. PMID: 16612252
-
Chromogranin A: a surprising link between granule biogenesis and hypertension.J Clin Invest. 2005 Jul;115(7):1711-3. doi: 10.1172/JCI25706. J Clin Invest. 2005. PMID: 16007250 Free PMC article. Review.
-
Hypertension from targeted ablation of chromogranin A can be rescued by the human ortholog.J Clin Invest. 2005 Jul;115(7):1942-52. doi: 10.1172/JCI24354. J Clin Invest. 2005. PMID: 16007257 Free PMC article.
-
Catecholamine storage vesicles and the metabolic syndrome: The role of the chromogranin A fragment pancreastatin.Diabetes Obes Metab. 2006 Nov;8(6):621-33. doi: 10.1111/j.1463-1326.2006.00575.x. Diabetes Obes Metab. 2006. PMID: 17026486 Free PMC article. Review.
-
The catecholamine release-inhibitory "catestatin" fragment of chromogranin a: naturally occurring human variants with different potencies for multiple chromaffin cell nicotinic cholinergic responses.Mol Pharmacol. 2004 Nov;66(5):1180-91. doi: 10.1124/mol.104.002139. Epub 2004 Aug 23. Mol Pharmacol. 2004. PMID: 15326220
Cited by
-
Role of Catestatin in the Cardiovascular System and Metabolic Disorders.Front Cardiovasc Med. 2022 May 19;9:909480. doi: 10.3389/fcvm.2022.909480. eCollection 2022. Front Cardiovasc Med. 2022. PMID: 35665253 Free PMC article. Review.
-
Single nucleotide polymorphism discovery and haplotype analysis of Ca2+-dependent K+ channel beta-1 subunit.Pharmacogenet Genomics. 2007 Apr;17(4):267-75. doi: 10.1097/FPC.0b013e3280105235. Pharmacogenet Genomics. 2007. PMID: 17496725 Free PMC article.
-
Catestatin induces glycogenesis by stimulating the phosphoinositide 3-kinase-AKT pathway.Acta Physiol (Oxf). 2022 May;235(1):e13775. doi: 10.1111/apha.13775. Epub 2022 Feb 4. Acta Physiol (Oxf). 2022. PMID: 34985191 Free PMC article.
-
Naturally occurring variants of the dysglycemic peptide pancreastatin: differential potencies for multiple cellular functions and structure-function correlation.J Biol Chem. 2014 Feb 14;289(7):4455-69. doi: 10.1074/jbc.M113.520916. Epub 2013 Dec 12. J Biol Chem. 2014. PMID: 24338022 Free PMC article. Clinical Trial.
-
An ancestral variant of Secretogranin II confers regulation by PHOX2 transcription factors and association with hypertension.Hum Mol Genet. 2007 Jul 15;16(14):1752-64. doi: 10.1093/hmg/ddm123. Epub 2007 Jun 21. Hum Mol Genet. 2007. PMID: 17584765 Free PMC article.
References
Electronic-Database Information
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CHGA) - PubMed
-
- Primer3, http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi (for design of PCR primers)
-
- Protein Data Bank, http://www.rcsb.org/pdb/ (for the NMR structure of catestatin, entry “1lv4”)
-
- SWISS-MODEL at ExPASy, http://www.expasy.org/swissmod/SWISS-MODEL.html
-
- UCSC Genome Bioinformatics, http://genome.ucsc.edu/
References
-
- Barbosa JA, Gill BM, Takiyyuddin MA, O’Connor DT (1991) Chromogranin A: posttranslational modifications in secretory granules. Endocrinology 128:174–190 - PubMed
-
- Cadman PE, Rao F, Mahata SK, O’Connor DT (2002) Studies of the dysglycemic peptide pancreastatin, using a human forearm model. Ann NY Acad Sci 971:528–529 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

