Null leukemia inhibitory factor receptor (LIFR) mutations in Stuve-Wiedemann/Schwartz-Jampel type 2 syndrome
- PMID: 14740318
- PMCID: PMC1181927
- DOI: 10.1086/381715
Null leukemia inhibitory factor receptor (LIFR) mutations in Stuve-Wiedemann/Schwartz-Jampel type 2 syndrome
Abstract
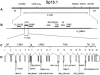
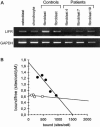
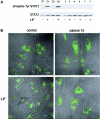
Stuve-Wiedemann syndrome (SWS) is a severe autosomal recessive condition characterized by bowing of the long bones, with cortical thickening, flared metaphyses with coarsened trabecular pattern, camptodactyly, respiratory distress, feeding difficulties, and hyperthermic episodes responsible for early lethality. Clinical overlap with Schwartz-Jampel type 2 syndrome (SJS2) has suggested that SWS and SJS2 could be allelic disorders. Through studying a series of 19 families with SWS/SJS2, we have mapped the disease gene to chromosome 5p13.1 at locus D5S418 (Zmax=10.66 at theta =0) and have identified null mutations in the leukemia inhibitory factor receptor (LIFR or gp190 chain) gene. A total of 14 distinct mutations were identified in the 19 families. An identical frameshift insertion (653_654insT) was identified in families from the United Arab Emirates, suggesting a founder effect in that region. It is interesting that 12/14 mutations predicted premature termination of translation. Functional studies indicated that these mutations alter the stability of LIFR messenger RNA transcripts, resulting in the absence of the LIFR protein and in the impairment of the JAK/STAT3 signaling pathway in patient cells. We conclude, therefore, that SWS and SJS2 represent a single clinically and genetically homogeneous condition due to null mutations in the LIFR gene on chromosome 5p13.
Figures

References
Electronic-Database Information
-
- UCSC Genome Bioinformatics, http://genome.ucsc.edu/ (for the human genome working draft)
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for LIFR [accession number NM_002310])
-
- National Center for Biotechnology Information (NCBI), http://www.ncbi.nlm.nih.gov/
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SWS) - PubMed
References
-
- Allan EH, Hilton DJ, Brown MA, Evely RS, Yumita S, Metcalf D, Gough NM, Nicola NA, Martin TJ (1990) Osteoblasts display receptors for and responses to leukemia-inhibitory factor. J Cell Physiol 145:110–119 - PubMed
-
- Bitard J, Daburon S, Duplomb L, Blanchard F, Vuisio P, Jacques Y, Godard A, Health JK, Moreau JF, Taupin JL (2003) Mutations in the immunoglobulin-like domain of gp190, the leukemia inhibitory factor (LIF) receptor, increase or decrease its affinity for LIF. J Biol Chem 278:16253–16261 10.1074/jbc.M207193200 - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

