Matching strategies for genetic association studies in structured populations
- PMID: 14740319
- PMCID: PMC1181929
- DOI: 10.1086/381716
Matching strategies for genetic association studies in structured populations
Abstract
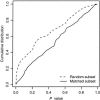
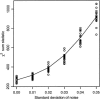
Association studies in populations that are genetically heterogeneous can yield large numbers of spurious associations if population subgroups are unequally represented among cases and controls. This problem is particularly acute for studies involving pooled genotyping of very large numbers of single-nucleotide-polymorphism (SNP) markers, because most methods for analysis of association in structured populations require individual genotyping data. In this study, we present several strategies for matching case and control pools to have similar genetic compositions, based on ancestry information inferred from genotype data for approximately 300 SNPs tiled on an oligonucleotide-based genotyping array. We also discuss methods for measuring the impact of population stratification on an association study. Results for an admixed population and a phenotype strongly confounded with ancestry show that these simple matching strategies can effectively mitigate the impact of population stratification.
Figures



References
Electronic-Database Information
-
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/ (for ss12673803–ss12674077)
-
- Genetic Epidemiology Group Web Site, http://www.lshtm.ac.uk/eu/genetics/ (for ADMIXMAP software)
-
- NCBI BLAST, http://www.ncbi.nlm.nih.gov/BLAST/ (for BLAST search engine)
-
- NCBI Home Page, http://www.ncbi.nlm.nih.gov/
-
- Pritchard Lab, http://pritch.bsd.uchicago.edu/ (for the structure program)
References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

