Mycobacterium bovis BCG cell wall-specific differentially expressed genes identified by differential display and cDNA subtraction in human macrophages
- PMID: 14742539
- PMCID: PMC321570
- DOI: 10.1128/IAI.72.2.937-948.2004
Mycobacterium bovis BCG cell wall-specific differentially expressed genes identified by differential display and cDNA subtraction in human macrophages
Abstract
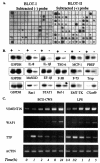
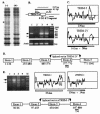
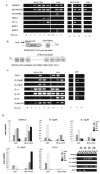
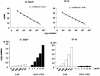
We have analyzed the gene expression profile of monocytes in response to a highly purified cell wall fraction of Mycobacterium bovis BCG, a clinically approved adjuvant known as BCG cell wall skeleton (BCG-CWS). It is composed of mycolic acid, arabinogalactan, and peptidoglycan and confers Toll-like receptor 2 (TLR2)- and TLR4-dependent signaling that induces monocytes to differentiate into antigen-presenting cells (APCs). Here we report differential gene expression analysis with BCG-CWS-stimulated versus nonstimulated monocytes. BCG-CWS exerted massive induction of genes regulated by TLR signaling. Marked gene regulatory characteristics in BCG-CWS-stimulated cells compared to lipopolysaccharide (LPS)-stimulated cells follow. (i) Spliced mRNAs encoding soluble forms of TREM-1 and TREM-2 (recently discovered inflammatory-signal-amplifying receptors) were regulated by BCG-CWS, resulting in their differential expression. (ii) The genes for zinc-iron transporter protein (ZIP)-like family proteins HKE-1 and LIV-1 were induced exclusively by BCG-CWS. (iii) Interleukin-23 (IL-23), rather than IL-12p70, was induced by BCG-CWS, while interferon-inducible genes were induced only by LPS. By Northern and reverse transcription-PCR analyses, we confirmed the differential expression of more than 30 BCG-CWS regulatory genes, and their expression was compared with that of LPS and other known TLR ligands. A battery of genes responded rapidly and for a short time to LPS but for a long time to BCG-CWS. Structural analysis of the identified novel or hypothetical proteins revealed that some are potential candidates as signaling mediators or transcriptional regulators. Hence, BCG-CWS may profoundly modulate APC responses in a way distinct from that of LPS, leading to possible advantages for its adjuvant-active therapeutic potential.
Figures



References
-
- Akazawa, T., H. Masuda, Y. Saeki, M. Matsumoto, K. Takeda, S. Akira, I. Azuma, K. Toyoshima, and T. Seya. Adjuvant-mediated tumor regression and tumor-specific CTL induction are impaired in MyD88-deficient mice. Cancer Res., in press. - PubMed
-
- Akira, S., K. Takeda, and T. Kaisho. 2001. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat. Immunol. 2:675-680. - PubMed
-
- Angell, J. E., D. J. Lindner, P. S. Shapiro, E. R. Hofmann, and D. V. Kalvakolanu. 2000. Identification of GRIM-19, a novel cell death-regulatory gene induced by the interferon-beta and retinoic acid combination, using a genetic approach. J. Biol. Chem. 275:33416-33426. - PubMed
-
- Azuma, I., and T. Seya. 2001. Development of immunoadjuvants for immunotherapy of cancer. Int. Immunopharmacol. 1:1249-1259. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

