Phylogenetic analysis of Sec7-domain-containing Arf nucleotide exchangers
- PMID: 14742722
- PMCID: PMC379250
- DOI: 10.1091/mbc.e03-06-0443
Phylogenetic analysis of Sec7-domain-containing Arf nucleotide exchangers
Abstract
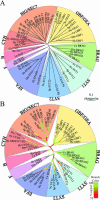
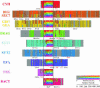
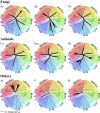
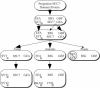
The eukaryotic family of ADP-ribosylation factor (Arf) GTPases plays a key role in the regulation of protein trafficking, and guanine-nucleotide exchange is crucial for Arf function. Exchange is stimulated by members of another family of proteins characterized by a 200-amino acid Sec7 domain, which alone is sufficient to catalyze exchange on Arf. Here, we analyzed the phylogeny of Sec7-domain-containing proteins in seven model organisms, representing fungi, plants, and animals. The phylogenetic tree has seven main groups, of which two include members from all seven model systems. Three groups are specific for animals, whereas two are specific for fungi. Based on this grouping, we propose a phylogenetically consistent set of names for members of the Sec7-domain family. Each group, except for one, contains proteins with known Arf exchange activity, implying that all members of this family have this activity. Contrary to the current convention, the sensitivity of Arf exchange activity to the inhibitor brefeldin A probably cannot be predicted by group membership. Multiple alignment reveals group-specific domains outside the Sec7 domain and a set of highly conserved amino acids within it. Determination of the importance of these conserved elements in Arf exchange activity and other cellular functions is now possible.
Figures

References
-
- Antonny, B., Beraud-Dufour, S., Chardin, P., and Chabre, M. (1997). N-terminal hydrophobic residues of the G-protein ADP-ribosylation factor-1 insert into membrane phospholipids upon GDP to GTP exchange. Biochemistry 36, 4675-4684. - PubMed
-
- Barlowe, C., and Schekman, R. (1993). SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature 365, 347-349. - PubMed
-
- Baumgartner, F., Wiek, S., Paprotka, K., Zauner, S., and Lingelbach, K. (2001). A point mutation in an unusual Sec7 domain is linked to brefeldin A resistance in a Plasmodium falciparum line generated by drug selection. Mol. Microbiol. 41, 1151-1158. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

