Rapid detection of single nucleotide polymorphisms associated with spinal muscular atrophy by use of a reusable fibre-optic biosensor
- PMID: 14742865
- PMCID: PMC373370
- DOI: 10.1093/nar/gnh013
Rapid detection of single nucleotide polymorphisms associated with spinal muscular atrophy by use of a reusable fibre-optic biosensor
Abstract
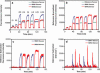
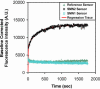
Rapid (<2 min) and quantitative genotyping for single nucleotide polymorphisms (SNPs) associated with spinal muscular atrophy was done using a reusable (approximately 80 cycles of application) fibre-optic biosensor over a clinically relevant range (0-4 gene copies). Sensors were functionalized with oligonucleotide probes immobilized at high density (approximately 7 pmol/cm2) to impart enhanced selectivity for SNP discrimination and used in a total internal reflection fluorescence detection motif to detect 202 bp PCR amplicons from patient samples. Real-time detection may be done over a range of ionic strength conditions (0.1-1.0 M) without stringency rinsing to remove non-selectively bound materials and without loss of selectivity, permitting a means for facile sample preparation. By using the time-derivative of fluorescence intensity as the analytical parameter, linearity of response may be maintained while allowing for significant reductions in analysis time (10-100-fold), permitting for the completion of measurements in under 1 min.
Figures




References
-
- Schena M. (1999) DNA Microarrays: A Practical Approach. Oxford University Press, New York, NY.
-
- Taton T.A., Mirkin,C.A. and Letsinger,R.L. (2000) Scanometric DNA array detection with nanoparticle probes. Science, 289, 1757–1760. - PubMed
-
- Yu C.J., Wan,Y., Yowanto,H., Li,J., Tao,C., James,M.D., Tan,C.L., Blackburn,G.F. and Meade,T.J. (2001) Electronic detection of single-base mismatches in DNA with ferrocene-modified probes. J. Am. Chem. Soc., 123, 11155–11161. - PubMed
-
- Duggan D.J., Bittner,M., Chen,Y., Meltzer,P. and Trent,J.M. (1999) Expression profiling using cDNA microarrays. Nature Genet., 21, 10–14. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

