Ovca1 regulates cell proliferation, embryonic development, and tumorigenesis
- PMID: 14744934
- PMCID: PMC338284
- DOI: 10.1101/gad.1162204
Ovca1 regulates cell proliferation, embryonic development, and tumorigenesis
Abstract
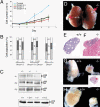
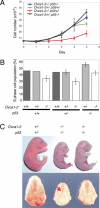
Loss of OVCA1/DPH2L1 correlates with ovarian and breast cancer. To study its in vivo role, we generated Ovca1 mutant alleles in mice. Ovca1 heterozygotes spontaneously develop cancer. Ovca1 mutant mice die during embryonic development and at birth with developmental delay and defects in multiple organ systems. Cell proliferation defects were observed in Ovca1 mutant mouse embryonic fibroblasts (MEFs). p53 deficiency can rescue these Ovca1 mutant MEF proliferation defects and partially rescue Ovca1 mutant embryonic phenotypes. Furthermore, Ovca1; p53 double heterozygotes developed tumors quicker than p53 heterozygotes and with an increased carcinoma incidence. Multiple tumor burden in Ovca1 heterozygotes that were also p53 deficient was significantly higher than in p53 homozygous mutants. These in vivo findings demonstrate that Ovca1 is a tumor suppressor that can modify p53-induced tumorigenesis and suggest that it acts as a positive regulator for cell cycle progression. The close linkage of OVCA1 and p53 on human Chromosome 17 suggests that coordinated loss may be an important mechanism for the evolution of ovarian, breast, and other tumor phenotypes.
Figures





Comment in
-
OVCA1: emerging as a bona fide tumor suppressor.Genes Dev. 2004 Feb 1;18(3):245-8. doi: 10.1101/gad.1184004. Genes Dev. 2004. PMID: 14871925 No abstract available.
References
-
- Atalay A., Crook, T., Ozturk, M., and Yulug, I.G. 2002. Identification of genes induced by BRCA1 in breast cancer cells. Biochem. Biophys. Res. Commun. 299: 839-846. - PubMed
-
- Bast R.C. and Mills, G.B. 2000. Alterations in oncogenes, tumor suppressor genes, and growth factors associated with epithelial ovarian cancers. In Ovarian cancer: Methods and protocols (ed. J.M.S. Bartlett), pp. 37-48. Humana Press, Totowa, NJ. - PubMed
-
- Blackburn A.C., Brown, J.S., Naber, S.P., Otis, C.N., Wood, J.T., and Jerry, D.J. 2003. BALB/c alleles for Prkdc and Cdkn2a interact to modify tumor susceptibility in Trp53+/- mice. Cancer Res. 63: 2364-2368. - PubMed
-
- Bruening W., Prowse, A.H., Schultz, D.C., Holgado-Madruga, M., Wong, A., and Godwin, A.K. 1999. Expression of OVCA1, a candidate tumor suppressor, is reduced in tumors and inhibits growth of ovarian cancer cells. Cancer Res. 59: 4973-4983. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
