Cassette-like variation of restriction enzyme genes in Escherichia coli C and relatives
- PMID: 14744977
- PMCID: PMC373321
- DOI: 10.1093/nar/gkh194
Cassette-like variation of restriction enzyme genes in Escherichia coli C and relatives
Abstract
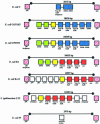
A surprising result of comparative bacterial genomics has been the large amount of DNA found to be present in one strain but not in another of the same species. We examine in detail one location where gene content varies extensively, the restriction cluster in Escherichia coli. This region is designated the Immigration Control Region (ICR) for the density and variability of restriction functions found there. To better define the boundaries of this variable locus, we determined the sequence of the region from a restrictionless strain, E.coli C. Here we compare the 13.7 kb E.coli C sequence spanning the site of the ICR with corresponding sequences from five E.coli strains and Salmonella typhimurium LT2. To discuss this variation, we adopt the term 'framework' to refer to genes that are stable components of genomes within related lineages, while 'migratory' genes are transient inhabitants of the genome. Strikingly, seven different migratory DNA segments, encoding different sets of genes and gene fragments, alternatively occupy a single well-defined location in the seven strains examined. The flanking framework genes, yjiS and yjiA, display approximately normal patterns of conservation. The patterns observed are consistent with the action of a site-specific recombinase. Since no nearby gene codes for a likely recombinase of known families, such a recombinase must be of a new family or unlinked.
Figures



References
-
- Raleigh E.A. (1992) Organization and function of the mcrBC genes of E.coli K-12. Mol. Microbiol., 6, 1079–1086. - PubMed
-
- Arber W. and Wauters-Willems,D. (1970) Host specificity of DNA produced by Escherichia coli. XII. The two restriction and modification systems of strain 15T-. Mol. Gen. Genet., 108, 203–217. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

