Protein structure prediction using sparse dipolar coupling data
- PMID: 14744980
- PMCID: PMC373331
- DOI: 10.1093/nar/gkh204
Protein structure prediction using sparse dipolar coupling data
Abstract
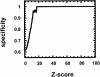
Residual dipolar coupling (RDC) represents one of the most exciting emerging NMR techniques for protein structure studies. However, solving a protein structure using RDC data alone is still a highly challenging problem. We report here a computer program, RDC-PROSPECT, for protein structure prediction based on a structural homolog or analog of the target protein in the Protein Data Bank (PDB), which best aligns with the (15)N-(1)H RDC data of the protein recorded in a single ordering medium. Since RDC-PROSPECT uses only RDC data and predicted secondary structure information, its performance is virtually independent of sequence similarity between a target protein and its structural homolog/analog, making it applicable to protein targets beyond the scope of current protein threading techniques. We have tested RDC-PROSPECT on all (15)N-(1)H RDC data (representing 43 proteins) deposited in the BioMagResBank (BMRB) database. The program correctly identified structural folds for 83.7% of the target proteins, and achieved an average alignment accuracy of 98.1% residues within a four-residue shift.
Figures

References
-
- Tjandra N. and Bax,A. (1997) Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science, 278, 1111–1114. - PubMed
-
- Tolman J.R. (2001) Dipolar couplings as a probe of molecular dynamics and structure in solution. Curr. Opin. Struct. Biol., 11, 532–539. - PubMed
-
- Prestegard J.H. (1998) New techniques in structural NMR–anisotropic interactions. Nature Struct. Biol., 5, 517–522. - PubMed
-
- Prestegard J.H., al-Hashimi,H.M. and Tolman,J.R. (2000) NMR structures of biomolecules using field oriented media and residual dipolar couplings. Q. Rev. Biophys., 33, 371–424. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

