Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis
- PMID: 14745009
- PMCID: PMC341741
- DOI: 10.1073/pnas.0307987100
Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis
Abstract
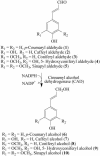
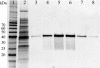
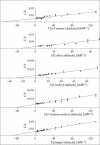
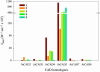
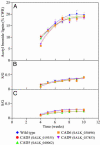
Of 17 genes annotated in the Arabidopsis genome database as cinnamyl alcohol dehydrogenase (CAD) homologues, an in silico analysis revealed that 8 genes were misannotated. Of the remaining nine, six were catalytically competent for NADPH-dependent reduction of p-coumaryl, caffeyl, coniferyl, 5-hydroxyconiferyl, and sinapyl aldehydes, whereas three displayed very low activity and only at very high substrate concentrations. Of the nine putative CADs, two (AtCAD5 and AtCAD4) had the highest activity and homology (approximately 83% similarity) relative to bona fide CADs from other species. AtCAD5 used all five substrates effectively, whereas AtCAD4 (of lower overall catalytic capacity) poorly used sinapyl aldehyde; the corresponding 270-fold decrease in k(enz) resulted from higher K(m) and lower k(cat) values, respectively. No CAD homologue displayed a specific requirement for sinapyl aldehyde, which was in direct contrast with unfounded claims for a so-called sinapyl alcohol dehydrogenase in angiosperms. AtCAD2, 3, as well as AtCAD7 and 8 (highest homology to sinapyl alcohol dehydrogenase) were catalytically less active overall by at least an order of magnitude, due to increased K(m) and lower k(cat) values. Accordingly, alternative and/or bifunctional metabolic roles of these proteins in plant defense cannot be ruled out. Comprehensive analyses of lignified tissues of various Arabidopsis knockout mutants (for AtCAD5, 6, and 9) at different stages of growth/development indicated the presence of functionally redundant CAD metabolic networks. Moreover, disruption of AtCAD5 expression had only a small effect on either overall lignin amounts deposited, or on syringyl-guaiacyl compositions, despite being the most catalytically active form in vitro.
Figures

References
-
- Gross, G. G., Stöckigt, J., Mansell, R. L. & Zenk, M. H. (1973) FEBS Lett. 31, 283-286.
-
- Mansell, R. L., Gross, G. G., Stöckigt, J., Franke, H. & Zenk, M. H. (1974) Phytochemistry 13, 2427-2435.
-
- Anterola, A. M. & Lewis, N. G. (2002) Phytochemistry 61, 221-294. - PubMed
-
- Lewis, N. G., Davin, L. B. & Sarkanen, S. (1999) in Comprehensive Natural Products Chemistry, eds. Barton, D. H. R., Nakanishi, K. & Meth-Cohn, O. (Elsevier, London), Vol. 3, pp. 617-745.
-
- Lewis, N. G. & Davin, L. B. (1999) in Comprehensive Natural Products Chemistry, eds. Barton, D. H. R., Nakanishi, K. & Meth-Cohn, O. (Elsevier, London), Vol. 1, pp. 639-712.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

