The Drosophila gene Start1: a putative cholesterol transporter and key regulator of ecdysteroid synthesis
- PMID: 14745013
- PMCID: PMC341787
- DOI: 10.1073/pnas.0308212100
The Drosophila gene Start1: a putative cholesterol transporter and key regulator of ecdysteroid synthesis
Abstract
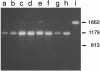
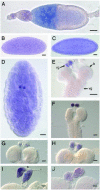
Human metastatic lymph node 64 (MLN64) is a transmembrane protein that shares homology with the cholesterol-binding vertebrate steroid acute regulatory protein (StAR)-related lipid transfer domain (START) and is involved in cholesterol traffic and steroid synthesis. We identified a Drosophila melanogaster gene whose putative protein product shows extensive homology with MLN64 and that we name Start1 (FlyBase CG3522). The putative Start1 protein, derived from Start1 cDNA sequences, contains an additional 122 aa of unknown function within the StAR-related lipid transfer domain. Similar inserts seem to exist in the Start1 homologues of Drosophila pseudoobscura and Anopheles gambiae, but not in the homologous protein of the urochordate Ciona intestinalis. Immunostaining using an insert-specific antibody confirms the presence of the insert in the cytoplasm. Whereas RT-PCR data indicate that Start1 is expressed ubiquitously, RNA in situ hybridizations demonstrate its overexpression in prothoracic gland cells, where ecdysteroids are synthesized from cholesterol. Transcripts of Start1 are detectable in embryonic ring gland progenitor cells and are abundant in prothoracic glands of larvae showing wave-like expression during larval stages. In adults, Start1 is expressed in nurse cells of the ovary. These observations are consistent with the assumption that Start1 plays a key role in the regulation of ecdysteroid synthesis. Vice versa, the expression of Start1 itself seems to depend on ecdysone, as in the ecdysone-deficient mutant ecd-1, Start1 expression is severely reduced.
Figures



References
-
- Becker, H. J. (1962) Chromosoma 13, 341-384.
-
- Thummel, C. S. (2002) Insect Biochem. Mol. Biol. 32, 113-120. - PubMed
-
- Siegmund, T. & Korge, G. (2001) J. Comp. Neurol. 431, 481-491. - PubMed
-
- Gilbert, L. I., Rybczynski, R. & Warren, J. T. (2002) Annu. Rev. Entomol. 47, 883-916. - PubMed
-
- Nagasawa, H., Guo, F., Zhong, X. C., Xia, B. Y., Wang, Z. S., Qui, X. J., Wei, D. Y., Chen, E. I., Wang, J. Z., Suzuki, A., et al. (1980) Sci. Sin. Ser. A 23, 1053-1060. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

