Evolution of the MAT locus and its Ho endonuclease in yeast species
- PMID: 14745027
- PMCID: PMC341799
- DOI: 10.1073/pnas.0304170101
Evolution of the MAT locus and its Ho endonuclease in yeast species
Abstract
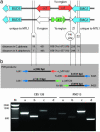
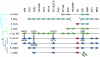
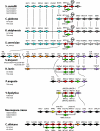
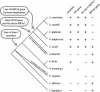
The genetics of the mating-type (MAT) locus have been studied extensively in Saccharomyces cerevisiae, but relatively little is known about how this complex system evolved. We compared the organization of MAT and mating-type-like (MTL) loci in nine species spanning the hemiascomycete phylogenetic tree. We inferred that the system evolved in a two-step process in which silent HMR/HML cassettes appeared, followed by acquisition of the Ho endonuclease from a mobile genetic element. Ho-mediated switching between an active MAT locus and silent cassettes exists only in the Saccharomyces sensu stricto group and their closest relatives: Candida glabrata, Kluyveromyces delphensis, and Saccharomyces castellii. We identified C. glabrata MTL1 as the ortholog of the MAT locus of K. delphensis and show that switching between C. glabrata MTL1a and MTL1alpha genotypes occurs in vivo. The more distantly related species Kluyveromyces lactis has silent cassettes but switches mating type without the aid of Ho endonuclease. Very distantly related species such as Candida albicans and Yarrowia lipolytica do not have silent cassettes. In Pichia angusta, a homothallic species, we found MATalpha2, MATalpha1, and MATa1 genes adjacent to each other on the same chromosome. Although some continuity in the chromosomal location of the MAT locus can be traced throughout hemiascomycete evolution and even to Neurospora, the gene content of the locus has changed with the loss of an HMG domain gene (MATa2) from the MATa idiomorph shortly after HO was recruited.
Figures
References
-
- Metzenberg, R. L. & Glass, N. L. (1990) BioEssays 12, 53-59. - PubMed
-
- Oshima, Y. (1993) in The Early Days of Yeast Genetics, eds. Hall, M. N. & Linder, P. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 291-304.
-
- Hicks, J. B., Strathern, J. N. & Herskowitz, I. (1977) in DNA Insertion Elements, Plasmids and Episomes, eds. Bukhari, A., Shapiro, J. & Adhya, S. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 457-462.
-
- Herskowitz, I., Rine, J. & Strathern, J. N. (1992) in The Molecular and Cellular Biology of the Yeast Saccharomyces, eds. Jones, E. W., Pringle, J. R. & Broach, J. R. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 583-656.
-
- Haber, J. E. (1998) Annu. Rev. Genet. 32, 561-599. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

