Analysis of ataxia-telangiectasia mutated (ATM)- and Nijmegen breakage syndrome (NBS)-regulated gene expression patterns
- PMID: 14745549
- PMCID: PMC12161825
- DOI: 10.1007/s00432-003-0522-y
Analysis of ataxia-telangiectasia mutated (ATM)- and Nijmegen breakage syndrome (NBS)-regulated gene expression patterns
Abstract
Purpose: Ataxia-telangiectasia (A-T) is a progressive, degenerative, complex autosomal recessive disease characterized by cerebellar degeneration, immunodeficiency, premature aging, radiosensitivity, and a predisposition to cancer. Mutations in the ataxia-telangiectasia mutated (atm) gene, which phosphorylates downstream effector proteins, are linked to A-T. One of the proteins phosphorylated by the ATM protein is Nijmegen Breakage Syndrome protein (NBS, p95/nibrin), which was recently shown to be encoded by a gene mutated in the Nijmegen breakage syndrome (nbs), an autosomal recessive disease with a phenotype virtually similar to that of A-T. The similarities in the clinical and cellular features of NBS and A-T have led us to hypothesize that the two corresponding gene products may function in similar ways in the cellular signaling pathway. Thus, we sought to identify genes whose expression is mediated by the atm and nbs gene products.
Material and methods: To identify genes, we performed an analysis of oligonucleotide microarrays using the appropriate cell lines, isogenic A-T (ATM-) and control cells (ATM+), and isogenic NBS (NBS-) and control cells (NBS+).
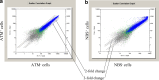
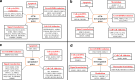
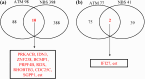
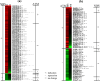
Results: We examined genes regulated by ATM and NBS, respectively. To determine the effect of ATM and NBS on gene expression in detail, we classified these genes into different functional categories, including those involved in apoptosis, cell cycle/DNA replication, growth/differentiation, signal transduction, cell-cell adhesion, and metabolism. In addition, we compared the genes regulated by the ATM and NBS to determine the relationship of their signaling pathways and to better understand their functional relationship.
Conclusions: We found that, while ATM and NBS regulate several genes in common, both of these proteins also have distinct patterns of gene regulation, findings consistent with the functional overlap and distinctiveness of these two conditions. Due to the role of ATM and NBS in tumor suppression and the response to chemotherapy and radiotherapy, these findings may assist in the development of a more rational approach to cancer treatment, as well as a better understanding of tumorigenesis.
Figures

References
-
- Blasina A, Price BD, Turenne GA, McGowan CH (1999) Caffeine inhibits the checkpoint kinase ATM. Curr Biol 9:1135–1138 - PubMed
-
- Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR 3rd, Hays L, Morgan WF, Petrini JH (1998) The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93:477–486 - PubMed
-
- Cortez D, Guntuku S, Qin J, Elledge SJ (2001) ATR and ATRIP: Partners in checkpoint signaling. Science 294:1713–1716 - PubMed
-
- Croxal JD, Newman SP, Choudhury Q, Flower RJ (1996) The concerted regulation of cPLA2, COX2, and lipocortin 1 expression by IL-1 beta in A 549 cells. Biochem Biophys Res Commun 220:491–495 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

