Activation of H-Ras in the endoplasmic reticulum by the RasGRF family guanine nucleotide exchange factors
- PMID: 14749369
- PMCID: PMC344182
- DOI: 10.1128/MCB.24.4.1516-1530.2004
Activation of H-Ras in the endoplasmic reticulum by the RasGRF family guanine nucleotide exchange factors
Abstract
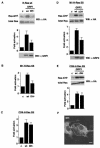
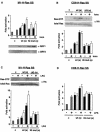
Recent findings indicate that in addition to its location in the peripheral plasma membrane, H-Ras is found in endomembranes like the endoplasmic reticulum and the Golgi complex. In these locations H-Ras is functional and can efficiently engage downstream effectors, but little is known about how its activation is regulated in these environments. Here we show that the RasGRF family exchange factors, both endogenous and ectopically expressed, are present in the endoplasmic reticulum but not in the Golgi complex. With the aid of H-Ras constructs specifically tethered to the plasma membrane, endoplasmic reticulum, and Golgi complex, we demonstrate that RasGRF1 and RasGRF2 can activate plasma membrane and reticular, but not Golgi-associated, H-Ras. We also show that RasGRF DH domain is required for the activation of H-Ras in the endoplasmic reticulum but not in the plasma membrane. Furthermore, we demonstrate that RasGRF mediation favors the activation of reticular H-Ras by lysophosphatidic acid treatment whereas plasma membrane H-Ras is made more responsive to stimulation by ionomycin. Overall, our results provide the initial insights into the regulation of H-Ras activation in the endoplasmic reticulum.
Figures
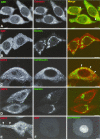
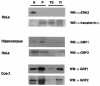
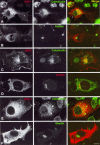
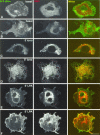


References
-
- Ajenjo, N., D. S. Aaronson, E. Ceballos, C. Richard, J. León, and P. Crespo. 2000. Myeloid leukemia cell growth and differentiation are independent of mitogen-activated protein kinases ERK1/2 activation. J. Biol. Chem 275:7189-7197. - PubMed
-
- Arozarena, I., D. S. Aaronson, D. Matallanas, V. Sanz, N. Ajenjo, S. Tenbaum, H. Teramoto, T. Ighishi, J. C. Zabala, J. S. Gutkind, and P. Crespo. 2000. The Rho family GTPase Cdc42 regulates the activation of Ras/MAPK pathway by the exchange factor Ras-GRF. J. Biol. Chem. 275:26441-26448. - PubMed
-
- Arozarena, I., D. Matallanas, and P. Crespo. 2001. Maintenance of Cdc42 GDP-bound state by Rho-GDI inhibits MAP kinase activation by the exchange factor Ras-GRF. J. Biol. Chem. 276:21878-21884. - PubMed
-
- Berridge, M. J., M. D. Bootman, and H. L. Roderick. 2003. Calcium signaling: dynamics homeostasis and remodeling. Nat. Rev. Mol. Cell Biol. 4:517-529. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
