Negative control of the Myc protein by the stress-responsive kinase Pak2
- PMID: 14749374
- PMCID: PMC344192
- DOI: 10.1128/MCB.24.4.1582-1594.2004
Negative control of the Myc protein by the stress-responsive kinase Pak2
Abstract
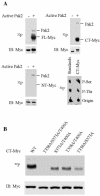
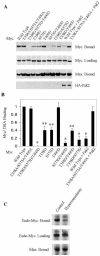
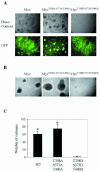
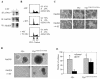
Pak2 is a serine/threonine kinase that participates in the cellular response to stress. Among the potential substrates for Pak2 is the protein Myc, encoded by the proto-oncogene MYC. Here we demonstrate that Pak2 phosphorylates Myc at three sites (T358, S373, and T400) and affects Myc functions both in vitro and in vivo. Phosphorylation at all three residues reduces the binding of Myc to DNA, either by blocking the requisite dimerization with Max (through phosphorylation at S373 and T400) or by interfering directly with binding to DNA (through phosphorylation at T358). Phosphorylation by Pak2 inhibits the ability of Myc to activate transcription, to sustain cellular proliferation, to transform NIH 3T3 cells in culture, and to elicit apoptosis on serum withdrawal. These results indicate that Pak2 is a negative regulator of Myc, suggest that inhibition of Myc plays a role in the cellular response to stress, and raise the possibility that Pak2 may be the product of a tumor suppressor gene.
Figures





References
-
- Alarcon-Vargas, D., W. P. Tansey, and Z. Ronai. 2002. Regulation of c-myc stability by selective stress conditions and by MEKK1 requires aa 127-189 of c-myc. Oncogene 21:4384-4391. - PubMed
-
- Alitalo, K., G. Ramsay, J. M. Bishop, S. O. Pfeifer, W. W. Colby, and A. D. Levinson. 1983. Identification of nuclear proteins encoded by viral and cellular myc oncogenes. Nature 306:274-277. - PubMed
-
- Amati, B., K. Alevizopoulos, and J. Vlach. 1998. Myc and the cell cycle. Front. Biosci. 3:D250-D268. - PubMed
-
- Axelson, H., M. Henriksson, Y. Wang, K. P. Magnusson, and G. Klein. 1995. The amino-terminal phosphorylation sites of C-MYC are frequently mutated in Burkitt's lymphoma lines but not in mouse plasmacytomas and rat immunocytomas. Eur. J. Cancer 31A:2099-2104. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
