Dnmt1 expression in pre- and postimplantation embryogenesis and the maintenance of IAP silencing
- PMID: 14749379
- PMCID: PMC344181
- DOI: 10.1128/MCB.24.4.1640-1648.2004
Dnmt1 expression in pre- and postimplantation embryogenesis and the maintenance of IAP silencing
Abstract
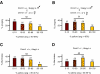
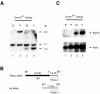
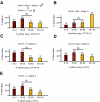
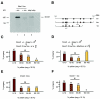
The methylation of intracisternal A-type particle (IAP) sequences is maintained during mouse embryogenesis. Methylation suppresses IAP expression and the potential for mutagenesis by retrotransposition, but it is not clear how methylation of these elements is maintained during the embryonic stages when the bulk of the genome is being demethylated. It has been suggested that the high levels of DNA methyltransferase-1 (Dnmt1) present during cleavage could be important for keeping IAPs methylated. To test this hypothesis, we combined mutant alleles of Dnmt1 with an agouti allele (A(iapy)), which provided a coat color readout for the methylation status of the IAP insertion in the agouti locus. We found that reduction in Dnmt1 levels directly impacted methylation at this locus, leading to stable transcriptional activation of the agouti gene in the adult. Specifically, the short maternal Dnmt1 protein was important in maintaining methylation at the A(iapy) locus in cleavage embryos, whereas the longer Dnmt1 isoform found in somatic cells was important in maintaining IAP methylation during the postimplantation stage. These results underscore the importance of maintaining proper maintenance of methylation patterns during gestation and suggest that interference with this process may stably affect gene expression patterns in the adult and may have profound phenotypic consequences.
Figures

References
-
- Bourc'his, D., G. L. Xu, C. S. Lin, B. Bollman, and T. H. Bestor. 2001. Dnmt3L and the establishment of maternal genomic imprints. Science 294:2536-2539. - PubMed
-
- Carlson, L. L., A. W. Page, and T. H. Bestor. 1992. Properties and localization of DNA methyltransferase in preimplantation mouse embryos: implications for genomic imprinting. Genes Dev. 6:2536-2541. - PubMed
-
- Chen, R. Z., U. Pettersson, C. Beard, L. Jackson-Grusby, and R. Jaenisch. 1998. DNA hypomethylation leads to elevated mutation rates. Nature 395:89-93. - PubMed
-
- Cooney, C. A., A. A. Dave, and G. L. Wolff. 2002. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J. Nutr. 132:2393S-2400S. - PubMed
-
- Duhl, D. M., H. Vrieling, K. A. Miller, G. L. Wolff, and G. S. Barsh. 1994. Neomorphic agouti mutations in obese yellow mice. Nat. Genet. 8:59-65. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
