U17/snR30 is a ubiquitous snoRNA with two conserved sequence motifs essential for 18S rRNA production
- PMID: 14749391
- PMCID: PMC344193
- DOI: 10.1128/MCB.24.4.1769-1778.2004
U17/snR30 is a ubiquitous snoRNA with two conserved sequence motifs essential for 18S rRNA production
Abstract
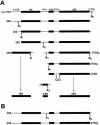
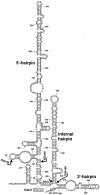
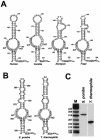
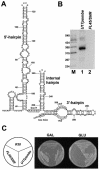
Saccharomyces cerevisiae snR30 is an essential box H/ACA small nucleolar RNA (snoRNA) required for the processing of 18S rRNA. Here, we show that the previously characterized human, reptilian, amphibian, and fish U17 snoRNAs represent the vertebrate homologues of yeast snR30. We also demonstrate that U17/snR30 is present in the fission yeast Schizosaccharomyces pombe and the unicellular ciliated protozoan Tetrahymena thermophila. Evolutionary comparison revealed that the 3'-terminal hairpins of U17/snR30 snoRNAs contain two highly conserved sequence motifs, the m1 (AUAUUCCUA) and m2 (AAACCAU) elements. Mutation analysis of yeast snR30 demonstrated that the m1 and m2 elements are essential for early cleavages of the 35S pre-rRNA and, consequently, for the production of mature 18S rRNA. The m1 and m2 motifs occupy the opposite strands of an internal loop structure, and they are located invariantly 7 nucleotides upstream from the ACA box of U17/snR30 snoRNAs. U17/snR30 is the first identified box H/ACA snoRNA that possesses an evolutionarily conserved role in the nucleolytic processing of eukaryotic pre-rRNA.
Figures



References
-
- Balakin, A. G., L. Smith, and M. J. Fournier. 1996. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell 86:823-834. - PubMed
-
- Borovjagin, A. V., and S. A. Gerbi. 2000. The spacing between functional cis-elements of U3 snoRNA is critical for rRNA processing. J. Mol. Biol. 300:57-74. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
