Repulsive guidance molecule (RGM) gene function is required for neural tube closure but not retinal topography in the mouse visual system
- PMID: 14749425
- PMCID: PMC6729817
- DOI: 10.1523/JNEUROSCI.4610-03.2004
Repulsive guidance molecule (RGM) gene function is required for neural tube closure but not retinal topography in the mouse visual system
Abstract
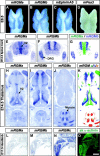
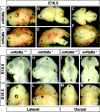
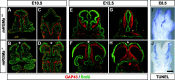
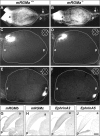
The establishment of topographic projections in the developing visual system depends on the spatially and temporally controlled expression of axon guidance molecules. In the developing chick tectum, the graded expression of the repulsive guidance molecule (RGM) has been proposed to be involved in controlling the topography of the retinal ganglion cell (RGC) axon termination zones along the anteroposterior axis of the tectum. We now show that there are three mouse proteins homologous to chick RGM displaying similar proteolytic processing but exhibiting differential cell-surface targeting by glycosyl phosphatidylinositol anchor addition. Two members of this gene family (mRGMa and mRGMb) are expressed in complementary patterns in the nervous system, and mRGMa is expressed prominently in the superior colliculus at the time of anteroposterior targeting of RGC axons. The third member of the family (mRGMc) is expressed almost exclusively in skeletal muscles. Functional studies in the mouse reveal a role for mRGMa in controlling cephalic neural tube closure, thus defining an unexpected role for mRGMa in early embryonic development. In contrast, mRGMa mutant mice did not exhibit defects in anteroposterior targeting of RGC axons to their stereotypic termination zones in the superior colliculus.
Figures




References
-
- Arber S, Han B, Mendelsohn M, Smith M, Jessell TM, Sockanathan S (1999) Requirement for the homeobox gene Hb9 in the consolidation of motor neuron identity. Neuron 23: 659–674. - PubMed
-
- Briscoe J, Pierani A, Jessell TM, Ericson J (2000) A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell 101: 435–445. - PubMed
-
- Brittis PA, Flanagan JG (2001) Nogo domains and a Nogo receptor: implications for axon regeneration. Neuron 30: 11–14. - PubMed
-
- Chen MS, Huber AB, van der Haar ME, Frank M, Schnell L, Spillmann AA, Christ F, Schwab ME (2000) Nogo-A is a myelin-associated neurite outgrowth inhibitor and an antigen for monoclonal antibody IN-1. Nature 403: 434–439. - PubMed
-
- Cheng HJ, Nakamoto M, Bergemann AD, Flanagan JG (1995) Complementary gradients in expression and binding of ELF-1 and Mek4 in development of the topographic retinotectal projection map. Cell 82: 371–381. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
