Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B
- PMID: 14752010
- PMCID: PMC324424
- DOI: 10.1101/gad.1149604
Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B
Abstract
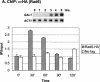
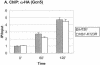
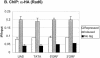
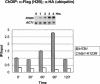
Covalent modifications of the histone N tails play important roles in eukaryotic gene expression. Histone acetylation, in particular, is required for the activation of a subset of eukaryotic genes through the targeted recruitment of histone acetyltransferases. We have reported that a histone C tail modification, ubiquitylation of H2B, is required for optimal expression of several inducible yeast genes, consistent with a role in transcriptional activation. H2B was shown to be ubiquitylated and then deubiquitylated at the GAL1 core promoter following galactose induction. We now show that the Rad6 protein, which catalyzes monoubiquitylation of H2B, is transiently associated with the GAL1 promoter upon gene activation, and that the period of its association temporally overlaps with the period of H2B ubiquitylation. Rad6 promoter association depends on the Gal4 activator and the Rad6-associated E3 ligase, Bre1, but is independent of the histone acetyltransferase, Gcn5. The SAGA complex, which contains a ubiquitin protease that targets H2B for deubiquitylation, is recruited to the GAL1 promoter in the absence of H2B ubiquitylation. The data suggest that Rad6 and SAGA function independently during galactose induction, and that the staged recruitment of these two factors to the GAL1 promoter regulates the ubiquitylation and deubiquitylation of H2B. We additionally show that both Rad6 and ubiquitylated H2B are absent from two regions of transcriptionally silent chromatin but present at genes that are actively transcribed. Thus, like histone H3 lysine 4 and lysine 79 methylation, two modifications that it regulates, Rad6-directed H2B ubiquitylation defines regions of active chromatin.
Figures










Similar articles
-
Histone H2B ubiquitylation is associated with elongating RNA polymerase II.Mol Cell Biol. 2005 Jan;25(2):637-51. doi: 10.1128/MCB.25.2.637-651.2005. Mol Cell Biol. 2005. PMID: 15632065 Free PMC article.
-
Rad6-Bre1-mediated histone H2B ubiquitylation modulates the formation of double-strand breaks during meiosis.Proc Natl Acad Sci U S A. 2004 Aug 3;101(31):11380-5. doi: 10.1073/pnas.0400078101. Epub 2004 Jul 27. Proc Natl Acad Sci U S A. 2004. PMID: 15280549 Free PMC article.
-
H2B ubiquitylation and de-ubiquitylation in gene activation.Novartis Found Symp. 2004;259:63-73; discussion 73-7, 163-9. Novartis Found Symp. 2004. PMID: 15171247 Review.
-
Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8.Genes Dev. 2003 Nov 1;17(21):2648-63. doi: 10.1101/gad.1144003. Epub 2003 Oct 16. Genes Dev. 2003. PMID: 14563679 Free PMC article.
-
H2B ubiquitylation: the end is in sight.Biochim Biophys Acta. 2004 Mar 15;1677(1-3):74-8. doi: 10.1016/j.bbaexp.2003.10.013. Biochim Biophys Acta. 2004. PMID: 15020048 Review.
Cited by
-
Mechanism for epigenetic variegation of gene expression at yeast telomeric heterochromatin.Genes Dev. 2012 Nov 1;26(21):2443-55. doi: 10.1101/gad.201095.112. Genes Dev. 2012. PMID: 23124068 Free PMC article.
-
Histone H2B monoubiquitination facilitates the rapid modulation of gene expression during Arabidopsis photomorphogenesis.PLoS Genet. 2012;8(7):e1002825. doi: 10.1371/journal.pgen.1002825. Epub 2012 Jul 19. PLoS Genet. 2012. PMID: 22829781 Free PMC article.
-
Bre1a, a histone H2B ubiquitin ligase, regulates the cell cycle and differentiation of neural precursor cells.J Neurosci. 2014 Feb 19;34(8):3067-78. doi: 10.1523/JNEUROSCI.3832-13.2014. J Neurosci. 2014. PMID: 24553946 Free PMC article.
-
Post-Translational Modifications of Histones Are Versatile Regulators of Fungal Development and Secondary Metabolism.Toxins (Basel). 2022 Apr 29;14(5):317. doi: 10.3390/toxins14050317. Toxins (Basel). 2022. PMID: 35622565 Free PMC article. Review.
-
UBIQUITIN-SPECIFIC PROTEASE 26 is required for seed development and the repression of PHERES1 in Arabidopsis.Genetics. 2008 Sep;180(1):229-36. doi: 10.1534/genetics.108.091736. Epub 2008 Aug 24. Genetics. 2008. PMID: 18723879 Free PMC article.
References
-
- Barsoum J. and Varshavsky, A. 1985. Preferential localization of varient nucleosomes near the 5′ end of the mouse dihydrofolate reductase gene. J. Biol. Chem. 260: 7688-7697. - PubMed
-
- Belotserkovskaya R. and Berger, S.L. 1999. Interplay between chromatin modifying and remodeling complexes in transcriptional regulation. Crit. Rev. Eukaryot. Gene Expr. 9: 221-230. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
