Molecular dissection of the roles of nucleotide binding and hydrolysis in dynein's AAA domains in Saccharomyces cerevisiae
- PMID: 14755060
- PMCID: PMC341754
- DOI: 10.1073/pnas.2637011100
Molecular dissection of the roles of nucleotide binding and hydrolysis in dynein's AAA domains in Saccharomyces cerevisiae
Retraction in
-
Molecular dissection of the roles of nucleotide binding and hydrolysis in dynein's AAA domains in Saccharomyces cerevisiae.Proc Natl Acad Sci U S A. 2004 Sep 28;101(39):14305. doi: 10.1073/pnas.0406339101. Proc Natl Acad Sci U S A. 2004. PMID: 15693149 Free PMC article. No abstract available.
Abstract
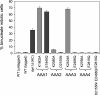
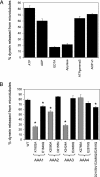
The motor protein cytoplasmic dynein is responsible for most of the minus-end-directed microtubule traffic within cells. Dynein contains four evolutionarily conserved AAA (ATPase associated with various cellular activities) domains that are thought to bind nucleotide; the role of nucleotide binding and hydrolysis in each of these four AAA domains has constituted an important and unresolved question in understanding dynein's mechanism. Using Saccharomyces cerevisiae cytoplasmic dynein as a model system, we mutagenized residues involved in nucleotide binding or hydrolysis in the four AAA domains and examined the ability of the mutant dyneins to mediate nuclear segregation in vivo and to bind microtubules in vitro. Our analysis shows that an AAA1 hydrolysis mutant blocks dynein function, whereas a triple AAA2/3/4 hydrolysis mutant does not, suggesting that nucleotide binding is required at only one site. We also show that nucleotide binding at AAA3, but not hydrolysis, is essential for motor activity in vivo and ATP-induced dissociation of dynein from microtubules, suggesting that this domain acts as a critical allosteric site. In contrast, mutations in AAA2 cause subtle defects in dynein function, whereas mutation in AAA4 produce no obvious defects. These results show that the four conserved dynein AAA domains have distinct functions in dynein's mechanochemical cycle.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

