Pathogenic profiles and molecular signatures of antinuclear autoantibodies rescued from NZM2410 lupus mice
- PMID: 14757744
- PMCID: PMC2211797
- DOI: 10.1084/jem.20030132
Pathogenic profiles and molecular signatures of antinuclear autoantibodies rescued from NZM2410 lupus mice
Abstract
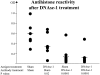
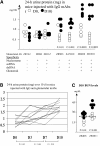
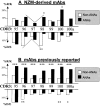
Two outstanding questions concerning antinuclear antibodies (ANAs) in lupus involve their pathogenic potential and their molecular signatures. To address these questions, a panel of 56 antinuclear and 47 nonnuclear binding monoclonal antibodies was rescued from four seropositive NZM2410 lupus mice. The monoclonals varied in their reactivity to nucleosomes, ssDNA, dsDNA, and glomerular substrate. A large fraction of the antibodies demonstrated apparent polyreactivity (to DNA, histones, and glomerular antigens) due to bound, DNase-1 sensitive nuclear antigenic bridges. Although nephrophilic immunoglobulin (Ig) M and IgG antibodies were the most pathogenic, the dsDNA-binding antibodies were modestly so; in contrast, antinucleosome antibodies were clearly not pathogenic. Compared with the nonnuclear antigen-binding monoclonal antibodies rescued from the same mice, ANAs exhibited increased utilization of VH5/7183 genes and highly cationic heavy chain (HC) CDR3 regions. Most intriguingly, the CDR3 regions of the ANAs exhibited alternating arginine/lysine peaks at H96, H98, and H100, with neutral troughs at H95, H97, and H99. To summarize, glomerular-binding anti-dsDNA antibodies appear to be the most pathogenic variety of lupus autoantibodies. The presence of an alternating charge pattern in their HC CDR3 regions appears to be a prominent hallmark of ANAs.
Figures


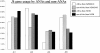


Similar articles
-
Utilization of the VH4-21 gene segment by anti-DNA antibodies from patients with systemic lupus erythematosus.J Autoimmun. 1993 Dec;6(6):809-25. doi: 10.1006/jaut.1993.1066. J Autoimmun. 1993. PMID: 8155258
-
Molecular characterization of anti-DNA antibodies induced in normal mice by immunization with bacterial DNA. Differences from spontaneous anti-DNA in the content and location of VH CDR3 arginines.J Immunol. 1993 Aug 1;151(3):1353-64. J Immunol. 1993. PMID: 8335932
-
Independently derived IgG anti-DNA autoantibodies from two lupus-prone mouse strains express a VH gene that is not present in most murine strains.J Immunol. 1993 Nov 1;151(9):4660-71. J Immunol. 1993. PMID: 8409427
-
Autoantibodies, lupus and the science of sabotage.Rheumatology (Oxford). 2004 Nov;43(11):1326-36. doi: 10.1093/rheumatology/keh354. Epub 2004 Aug 24. Rheumatology (Oxford). 2004. PMID: 15328419 Review.
-
[Role of the nucleosome in the physiopathology of systemic lupus erythematosus].Ann Med Interne (Paris). 2003 Feb;154(1):25-32. Ann Med Interne (Paris). 2003. PMID: 12746656 Review. French.
Cited by
-
Cellular and molecular pathogenesis of systemic lupus erythematosus: lessons from animal models.Arthritis Res Ther. 2011;13(5):241. doi: 10.1186/ar3465. Epub 2011 Sep 30. Arthritis Res Ther. 2011. PMID: 21989039 Free PMC article. Review.
-
What is damaging the kidney in lupus nephritis?Nat Rev Rheumatol. 2016 Mar;12(3):143-53. doi: 10.1038/nrrheum.2015.159. Epub 2015 Nov 19. Nat Rev Rheumatol. 2016. PMID: 26581344 Free PMC article. Review.
-
Anti-nuclear antibody reactivity in lupus may be partly hard-wired into the primary B-cell repertoire.Mol Immunol. 2009 Oct;46(16):3420-6. doi: 10.1016/j.molimm.2009.07.014. Epub 2009 Aug 21. Mol Immunol. 2009. PMID: 19699528 Free PMC article.
-
Kallikrein genes are associated with lupus and glomerular basement membrane-specific antibody-induced nephritis in mice and humans.J Clin Invest. 2009 Apr;119(4):911-23. doi: 10.1172/JCI36728. J Clin Invest. 2009. PMID: 19307730 Free PMC article.
-
The role of rearrangement at the second Ig heavy chain locus in maintaining B cell tolerance to DNA.J Immunol. 2008 Jun 1;180(11):7721-7. doi: 10.4049/jimmunol.180.11.7721. J Immunol. 2008. PMID: 18490776 Free PMC article.
References
-
- Kotzin, B.L. 1996. Systemic lupus erythematosus. Cell. 85:303–306. - PubMed
-
- Hahn, B.H. 1998. Antibodies to DNA. N. Engl. J. Med. 338:1359–1368. - PubMed
-
- Pisetsky, D.H. 2000. Anti-DNA and autoantibodies. Curr. Opin. Rheumatol. 12:364–368. - PubMed
-
- Lefkowith, J.B., and G.S. Gilkeson. 1996. Nephritogenic autoantibodies in lupus. Current concepts and continuing controversies. Arthritis Rheum. 39:894–903. - PubMed
-
- Vlahakos, D.V., M.H. Foster, S. Adams, M. Katz, A.A. Ucci, K.J. Barrett, S.K. Datta, and M.P. Madaio. 1992. Anti-DNA antibodies form immune deposits at distinct glomerular and vascular sites. Kidney Int. 41:1690–1700. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

