Architecture of the Bacteroides cellulosolvens cellulosome: description of a cell surface-anchoring scaffoldin and a family 48 cellulase
- PMID: 14761991
- PMCID: PMC344227
- DOI: 10.1128/JB.186.4.968-977.2004
Architecture of the Bacteroides cellulosolvens cellulosome: description of a cell surface-anchoring scaffoldin and a family 48 cellulase
Abstract
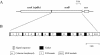
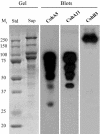
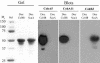
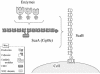
A large gene downstream of the primary Bacteroides cellulosolvens cellulosomal scaffoldin (cipBc, now renamed scaA) was sequenced. The gene, termed scaB, contained an N-terminal leader peptide followed by 10 type I cohesins, an "X" domain of unknown structure and function, and a C-terminal S-layer homology (SLH) surface-anchoring module. In addition, a previously identified gene in a different part of the genome, encoding for a dockerin-borne family 48 cellulosomal glycoside hydrolase (Cel48), was sequenced completely, and a putative cellulosome-related family 9 glycosyl hydrolase was detected. Recombinant fusion proteins, comprising dockerins derived from either the ScaA scaffoldin or Cel48, were overexpressed. Their interaction with ScaA and ScaB cohesins was examined by immunoassay. The results indicated that the ScaB type I cohesin of the new anchoring protein binds selectively to the ScaA dockerin, whereas the Cel48 dockerin binds specifically to the type II ScaA cohesin 5. Thus, by virtue of the 11 type II ScaA cohesins and the 10 type I ScaB cohesins, the relatively simple two-component cellulosome-integrating complex would potentially incorporate 110 enzyme molecules onto the cell surface via the ScaB SLH module. Compared to previously described cellulosome systems, the apparent roles of the B. cellulosolvens cohesins are reversed, in that the type II cohesins are located on the enzyme-binding primary scaffoldin, whereas the type I cohesins are located on the anchoring scaffoldin. The results underscore the extensive diversity in the supramolecular architecture of cellulosome systems in nature.
Figures




References
-
- Bagnara-Tardif, C., C. Gaudin, A. Belaich, P. Hoest, T. Citard, and J.-P. Belaich. 1992. Sequence analysis of a gene cluster encoding cellulases from Clostridium cellulolyticum. Gene 119:17-28. - PubMed
-
- Bayer, E. A., H. Chanzy, R. Lamed, and Y. Shoham. 1998. Cellulose, cellulases and cellulosomes. Curr. Opin. Struct. Biol. 8:548-557. - PubMed
-
- Bayer, E. A., E. Morag, and R. Lamed. 1994. The cellulosome: a treasure-trove for biotechnology. Trends Biotechnol. 12:378-386. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

