Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42
- PMID: 14762003
- PMCID: PMC344220
- DOI: 10.1128/JB.186.4.1084-1096.2004
Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42
Abstract
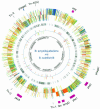
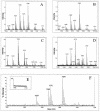
The environmental strain Bacillus amyloliquefaciens FZB42 promotes plant growth and suppresses plant pathogenic organisms present in the rhizosphere. We sampled sequenced the genome of FZB42 and identified 2,947 genes with >50% identity on the amino acid level to the corresponding genes of Bacillus subtilis 168. Six large gene clusters encoding nonribosomal peptide synthetases (NRPS) and polyketide synthases (PKS) occupied 7.5% of the whole genome. Two of the PKS and one of the NRPS encoding gene clusters were unique insertions in the FZB42 genome and are not present in B. subtilis 168. Matrix-assisted laser desorption ionization-time of flight mass spectrometry analysis revealed expression of the antibiotic lipopeptide products surfactin, fengycin, and bacillomycin D. The fengycin (fen) and the surfactin (srf) operons were organized and located as in B. subtilis 168. A large 37.2-kb antibiotic DNA island containing the bmy gene cluster was attributed to the biosynthesis of bacillomycin D. The bmy island was found inserted close to the fen operon. The responsibility of the bmy, fen, and srf gene clusters for the production of the corresponding secondary metabolites was demonstrated by cassette mutagenesis, which led to the loss of the ability to produce these peptides. Although these single mutants still largely retained their ability to control fungal spread, a double mutant lacking both bacillomycin D and fengycin was heavily impaired in its ability to inhibit growth of phytopathogenic fungi, suggesting that both lipopeptides act in a synergistic manner.
Figures




References
-
- Ceglowski, P., and J. C. Alonso. 1994. Gene organization of the Streptococcus pyogenes plasmid pDB101: sequence analysis of the ORF eta-copS region. Gene 145:33-39. - PubMed
-
- Challis, G. L., J. Ravel, and C. A. Townsend. 2000. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 7:211-224. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

