Mechanism of transcription activation at the comG promoter by the competence transcription factor ComK of Bacillus subtilis
- PMID: 14762007
- PMCID: PMC344208
- DOI: 10.1128/JB.186.4.1120-1128.2004
Mechanism of transcription activation at the comG promoter by the competence transcription factor ComK of Bacillus subtilis
Abstract
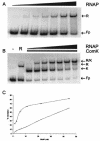
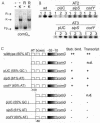
The development of genetic competence in Bacillus subtilis is regulated by a complex signal transduction cascade, which results in the synthesis of the competence transcription factor, encoded by comK. ComK is required for the transcription of the late competence genes that encode the DNA binding and uptake machinery and of genes required for homologous recombination. In vivo and in vitro experiments have shown that ComK is responsible for transcription activation at the comG promoter. In this study, we investigated the mechanism of this transcription activation. The intrinsic binding characteristics of RNA polymerase with and without ComK at the comG promoter were determined, demonstrating that ComK stabilizes the binding of RNA polymerase to the comG promoter. This stabilization probably occurs through interactions with the upstream DNA, since a deletion of the upstream DNA resulted in an almost complete abolishment of stabilization of RNA polymerase binding. Furthermore, a strong requirement for the presence of an extra AT box in addition to the common ComK-binding site was shown. In vitro transcription with B. subtilis RNA polymerase reconstituted with wild-type alpha-subunits and with C-terminal deletion mutants of the alpha-subunits was performed, demonstrating that these deletions do not abolish transcription activation by ComK. This indicates that ComK is not a type I activator. We also show that ComK is not required for open complex formation. A possible mechanism for transcription activation is proposed, implying that the major stimulatory effect of ComK is on binding of RNA polymerase.
Figures



Similar articles
-
A single, specific thymine mutation in the ComK-binding site severely decreases binding and transcription activation by the competence transcription factor ComK of Bacillus subtilis.J Bacteriol. 2007 Jul;189(13):4718-28. doi: 10.1128/JB.00281-07. Epub 2007 Apr 27. J Bacteriol. 2007. PMID: 17468244 Free PMC article.
-
The competence transcription factor of Bacillus subtilis recognizes short A/T-rich sequences arranged in a unique, flexible pattern along the DNA helix.Genes Dev. 1998 May 15;12(10):1539-50. doi: 10.1101/gad.12.10.1539. Genes Dev. 1998. PMID: 9585513 Free PMC article.
-
comK encodes the competence transcription factor, the key regulatory protein for competence development in Bacillus subtilis.Mol Microbiol. 1995 Feb;15(3):455-62. doi: 10.1111/j.1365-2958.1995.tb02259.x. Mol Microbiol. 1995. PMID: 7783616
-
Improving the predictive value of the competence transcription factor (ComK) binding site in Bacillus subtilis using a genomic approach.Nucleic Acids Res. 2002 Dec 15;30(24):5517-28. doi: 10.1093/nar/gkf698. Nucleic Acids Res. 2002. PMID: 12490720 Free PMC article.
-
Regulatory inputs for the synthesis of ComK, the competence transcription factor of Bacillus subtilis.Mol Microbiol. 1996 Aug;21(4):763-75. doi: 10.1046/j.1365-2958.1996.371407.x. Mol Microbiol. 1996. PMID: 8878039
Cited by
-
Crystal structures of two transcriptional regulators from Bacillus cereus define the conserved structural features of a PadR subfamily.PLoS One. 2012;7(11):e48015. doi: 10.1371/journal.pone.0048015. Epub 2012 Nov 26. PLoS One. 2012. PMID: 23189126 Free PMC article.
-
A single, specific thymine mutation in the ComK-binding site severely decreases binding and transcription activation by the competence transcription factor ComK of Bacillus subtilis.J Bacteriol. 2007 Jul;189(13):4718-28. doi: 10.1128/JB.00281-07. Epub 2007 Apr 27. J Bacteriol. 2007. PMID: 17468244 Free PMC article.
-
Functional analysis of the ComK protein of Bacillus coagulans.PLoS One. 2013;8(1):e53471. doi: 10.1371/journal.pone.0053471. Epub 2013 Jan 3. PLoS One. 2013. PMID: 23301076 Free PMC article.
-
Listeria monocytogenes σH Contributes to Expression of Competence Genes and Intracellular Growth.J Bacteriol. 2016 Mar 31;198(8):1207-17. doi: 10.1128/JB.00718-15. Print 2016 Apr. J Bacteriol. 2016. PMID: 26833412 Free PMC article.
-
Allostery through DNA drives phenotype switching.Nat Commun. 2021 May 20;12(1):2967. doi: 10.1038/s41467-021-23148-2. Nat Commun. 2021. PMID: 34016970 Free PMC article.
References
-
- Adhya, S., and S. Garges. 1990. Positive control. J. Biol. Chem. 265:10797-10800. - PubMed
-
- Adhya, S., M. Gottesman, S. Garges, and A. Oppenheim. 1993. Promoter resurrection by activators—a minireview. Gene 132:1-6. - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Current protocols in molecular biology. Green Publishing Associates and Wiley Interscience, New York, N.Y.
-
- Berka, R. M., J. Hahn, M. Albano, I. Draskovic, M. Persuh, X. Cui, A. Sioma, W. Widner, and D. Dubnau. 2002. Microarray analysis of the Bacillus subtilis K-state: genome-wide expression changes dependent on ComK. Mol. Microbiol. 43:1331-1345. - PubMed
-
- Borowiec, J. A., and J. D. Gralla. 1987. All three elements of the lac pS promoter mediate its transcriptional response to DNA supercoiling. J. Mol. Biol. 195:89-97. - PubMed

