A motif co-occurrence approach for genome-wide prediction of transcription-factor-binding sites in Escherichia coli
- PMID: 14762058
- PMCID: PMC327095
- DOI: 10.1101/gr.1448004
A motif co-occurrence approach for genome-wide prediction of transcription-factor-binding sites in Escherichia coli
Abstract
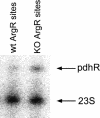
Various computational approaches have been developed for predicting cis-regulatory DNA elements in prokaryotic genomes. We describe a novel method for predicting transcription-factor-binding sites in Escherichia coli. Our method takes advantage of the principle that transcription factors frequently coregulate gene expression, but without requiring prior knowledge of which groups of genes are coregulated. Using position weight matrices for 49 known transcription factors, we examined spacings between pairs of matrix hits. These pairs were assigned probabilities according to the overrepresentation of their separation distance. The functions of many open reading frames (ORFs) downstream from predicted binding sites are unknown, and may correspond to novel regulon members. For five predictions, knockouts with mutated replacements of the predicted binding sites were created in E. coli MG1655. Quantitative real-time PCR (RT-PCR) indicates that for each of the knockouts, at least one gene immediately downstream exhibits a statistically significant change in mRNA expression. This approach may be useful in analyzing binding sites in a variety of organisms.
Figures

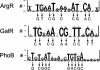
References
-
- Affymetrix, Inc. 2002. Affymetrix GeneChip Expression Analysis Technical Manual. Affymetrix, Inc., Santa Clara, CA.
-
- Bailey, T. and Elkan, C. 1995. The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 3: 21-29. - PubMed
-
- Berg, O. and von Hippel, P. 1987. Selection of DNA binding sites by regulatory proteins. Statistical-mechanical theory and application to operators and promoters. J. Mol. Biol. 193: 723-750. - PubMed
-
- Berman, B.P., Nibu, Y., Pfeiffer, B.D., Tomancak, P., Celniker, S.E., Levine, M., Rubin, G.M., and Eisen, M.B. 2002. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl. Acad. Sci. 99: 757-762. - PMC - PubMed
WEB SITE REFERENCES
-
- http://arep.med.harvard.edu/ecoli_matrices/spacing/spacing_predictions.html; Web site contains tab-delimited files containing predictions based on individual spacings, and separately based on spacing bins.
-
- http://arep.med.harvard.edu/labgc/pko3.html; Descriptions of the gene replacement vectors pKO3 and pKOV.
-
- http://twod.med.harvard.edu/labgc/estep/longPCR_protocol.html; Descriptions of the PCR conditions and protocols used in this project.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
