Activin regulation of the follicle-stimulating hormone beta-subunit gene involves Smads and the TALE homeodomain proteins Pbx1 and Prep1
- PMID: 14764653
- PMCID: PMC2935796
- DOI: 10.1210/me.2003-0442
Activin regulation of the follicle-stimulating hormone beta-subunit gene involves Smads and the TALE homeodomain proteins Pbx1 and Prep1
Abstract
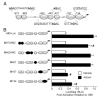
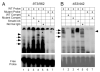
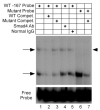
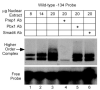
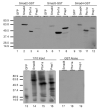
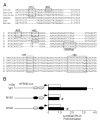
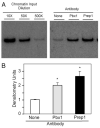
FSH is critical for normal reproductive function in both males and females. Activin, a member of the TGFbeta family of growth factors, is an important regulator of FSH expression, but little is known about the molecular mechanisms through which it acts. We used transient transfections into the immortalized gonadotrope cell line LbetaT2 to identify three regions (at -973/-962, -167, and -134) of the ovine FSH beta-subunit gene that are required for full activin response. All three regions contain homology to consensus binding sites for Smad proteins, the intracellular mediators of TGFbeta family signaling. Mutation of the distal site reduces activin responsiveness, whereas mutation of either proximal site profoundly disrupts activin regulation of the FSHbeta gene. These sites specifically bind LbetaT2 nuclear proteins in EMSAs, and the -973/-962 site binds Smad4 protein. Interestingly, the protein complex binding to the -134 site contains Smad4 in association with the homeodomain proteins Pbx1 and Prep1. Using glutathione S-transferase interaction assays, we demonstrate that Pbx1 and Prep1 interact with Smads 2 and 3 as well. The two proximal activin response elements are well conserved across species, and Pbx1 and Prep1 proteins bind to the mouse gene in vivo. Furthermore, mutation of either proximal site abrogates activin responsiveness of a mouse FSHbeta reporter gene as well, confirming their functional conservation. Our studies provide a basis for understanding activin regulation of FSHbeta gene expression and identify Pbx1 and Prep1 as Smad partners and novel mediators of activin action.
Figures


References
-
- Ling N, Ying S-Y, Ueno N, Shimasaki S, Esch F, Hotta M, Guillemin R. Pituitary FSH is released by a heterodimer of the β-subunits from the two forms of inhibin. Nature. 1986;321:779–782. - PubMed
-
- Carroll RS, Corrigan AZ, Gharib SD, Vale W, Chin WW. Inhibin, activin, and follistatin: regulation of follicle-stimulating hormone messenger ribonucleic acid levels. Mol Endocrinol. 1989;3:1969–1976. - PubMed
-
- Carroll RS, Kowash PM, Lofgren JA, Schwall RH, Chin WW. In vivo regulation of FSH synthesis by inhibin and activin. Endocrinology. 1991;129:3299–3304. - PubMed
-
- Matzuk MM, Kumar TR, Bradley A. Different phenotypes for mice deficient in either activins or activin receptor type II. Nature. 1995;374:356–560. - PubMed
-
- Attisano L, Wrana JL. Signal transduction by the TGF-β superfamily. Science. 2002;296:1646–1647. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 HD41301/HD/NICHD NIH HHS/United States
- T32 DK007044/DK/NIDDK NIH HHS/United States
- R01 HD020377/HD/NICHD NIH HHS/United States
- T32 GM08666/GM/NIGMS NIH HHS/United States
- T32 DK007541/DK/NIDDK NIH HHS/United States
- R37 HD20377/HD/NICHD NIH HHS/United States
- F32 HD041301/HD/NICHD NIH HHS/United States
- U54 HD012303/HD/NICHD NIH HHS/United States
- T32 DK07541/DK/NIDDK NIH HHS/United States
- R37 HD020377/HD/NICHD NIH HHS/United States
- T32 GM008666/GM/NIGMS NIH HHS/United States
- U54 HD12303/HD/NICHD NIH HHS/United States
- P50 HD012303/HD/NICHD NIH HHS/United States
- T32 DK07044/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

