Lipoic acid-dependent oxidative catabolism of alpha-keto acids in mitochondria provides evidence for branched-chain amino acid catabolism in Arabidopsis
- PMID: 14764908
- PMCID: PMC344558
- DOI: 10.1104/pp.103.035675
Lipoic acid-dependent oxidative catabolism of alpha-keto acids in mitochondria provides evidence for branched-chain amino acid catabolism in Arabidopsis
Abstract
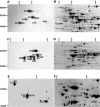
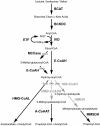
Lipoic acid-dependent pathways of alpha-keto acid oxidation by mitochondria were investigated in pea (Pisum sativum), rice (Oryza sativa), and Arabidopsis. Proteins containing covalently bound lipoic acid were identified on isoelectric focusing/sodium dodecyl sulfate-polyacrylamide gel electrophoresis separations of mitochondrial proteins by the use of antibodies raised to this cofactor. All these proteins were identified by tandem mass spectrometry. Lipoic acid-containing acyltransferases from pyruvate dehydrogenase complex and alpha-ketoglutarate dehydrogenase complex were identified from all three species. In addition, acyltransferases from the branched-chain dehydrogenase complex were identified in both Arabidopsis and rice mitochondria. The substrate-dependent reduction of NAD(+) was analyzed by spectrophotometry using specific alpha-keto acids. Pyruvate- and alpha-ketoglutarate-dependent reactions were measured in all three species. Activity of the branched-chain dehydrogenase complex was only measurable in Arabidopsis mitochondria using substrates that represented the alpha-keto acids derived by deamination of branched-chain amino acids (Val [valine], leucine, and isoleucine). The rate of branched-chain amino acid- and alpha-keto acid-dependent oxygen consumption by intact Arabidopsis mitochondria was highest with Val and the Val-derived alpha-keto acid, alpha-ketoisovaleric acid. Sequencing of peptides derived from trypsination of Arabidopsis mitochondrial proteins revealed the presence of many of the enzymes required for the oxidation of all three branched-chain amino acids. The potential role of branched-chain amino acid catabolism as an oxidative phosphorylation energy source or as a detoxification pathway during plant stress is discussed.
Figures
References
-
- Aubert S, Alban C, Bligny R, Douce R (1996) Induction of beta-methylcrotonyl-coenzyme A carboxylase in higher plant cells during carbohydrate starvation: evidence for a role of MCCase in leucine catabolism. FEBS Lett 383: 175–180 - PubMed
-
- Chew O, Whelan J (2003) Dual targeting ability of targeting signals is dependent on the nature of the mature protein. Funct Plant Biol 30: 805–812 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

