Mapping key functional sites within yeast TFIID
- PMID: 14765106
- PMCID: PMC381015
- DOI: 10.1038/sj.emboj.7600111
Mapping key functional sites within yeast TFIID
Abstract
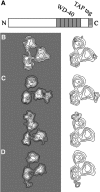
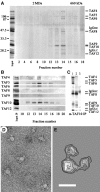
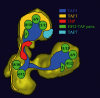
The transcription factor TFIID, composed of the TATA box-binding protein (TBP) and 14 TBP-associated factors (TAFs), plays a key role in the regulation of gene expression by RNA polymerase II. The structure of yeast TFIID, as determined by electron microscopy and digital image analysis, is formed by three lobes, labelled A-C, connected by thin linking domains. Immunomapping revealed that TFIID contains two copies of the WD-40 repeat-containing TAF5 and that TAF5 contributes to the linkers since its C- and N-termini were found in different lobes. This property was confirmed by the finding that a recombinant complex containing TAF5 complexed with six histone fold containing TAFs was able to form a trilobed structure. Moreover, the N-terminal domain of TAF1 was mapped in lobe C, whereas the histone acetyltransferase domain resides in lobe A along with TAF7. TBP was found in the linker domain between lobes A and C in a way that the N-terminal 100 residues of TAF1 are spanned over it. The implications of these data with regard to TFIID function are discussed.
Figures


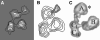
References
-
- Andel F, Ladurner AG, Inouye C, Tjian R, Nogales E (1999) Three-dimensional structure of the human TFIID–IIA–IIB complex. Science 286: 2153–2156 - PubMed
-
- Banik U, Beechem JM, Klebanow E, Schroeder S, Weil PA (2001) Fluorescence-based analyses of the effects of full-length recombinant TAF130p on the interaction of TATA box-binding protein with TATA box DNA. J Biol Chem 276: 49100–49109 - PubMed
-
- Brand M, Leurent C, Mallouh V, Tora L, Schultz P (1999) Three-dimensional structures of the TAFII-containing complexes TFIID and TFTC. Science 286: 2151–2153 - PubMed
-
- Buratowski S, Hahn S, Guarente L, Sharp PA (1989) Five intermediate complexes in transcription initiation by RNA polymerase II. Cell 56: 549–561 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

