Fission yeast Clp1p phosphatase affects G2/M transition and mitotic exit through Cdc25p inactivation
- PMID: 14765109
- PMCID: PMC381010
- DOI: 10.1038/sj.emboj.7600103
Fission yeast Clp1p phosphatase affects G2/M transition and mitotic exit through Cdc25p inactivation
Abstract
The Cdc14 family of phosphatases specifically reverses proline-directed phosphorylation events. In Saccharomyces cerevisiae, Cdc14p promotes Cdk1p inactivation at mitotic exit by reversing Cdk1p-dependent phosphorylations. Cdk1p is a proline-directed kinase whose activity is required in all eukaryotes for the transit into mitosis. At mitotic commitment, Cdk1p participates in its own regulation by activating the mitotic inducing phosphatase, Cdc25p, and inhibiting the opposing kinase, Wee1p. We have investigated the ability of Schizosaccharomyces pombe Clp1p, a Cdc14p homolog, to disrupt this auto-amplification loop. We show here that Clp1p is required to dephosphorylate, destabilize, and inactivate Cdc25p at the end of mitosis. Clp1p promotes recognition of Cdc25p by the anaphase-promoting complex/cyclosome, an E3 ubiquitin ligase. Failure to inactivate and destabilize Cdc25p in late mitosis delays progression through anaphase, interferes with septation initiation network signaling, and additionally advances the commitment to mitotic entry in the next cycle. This may be a widely conserved mechanism whereby Cdc14 proteins contribute to Cdk1p inactivation.
Figures

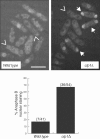





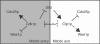
References
-
- Aligue R, Wu L, Russell P (1997) Regulation of Schizosaccharomyces pombe Wee1 tyrosine kinase. J Biol Chem 272: 13320–13325 - PubMed
-
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A 3rd, Steever AB, Wach A, Philippsen P, Pringle JR (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951 - PubMed
-
- Bardin AJ, Amon A (2001) Men and sin: what's the difference? Nat Rev Mol Cell Biol 2: 815–826 - PubMed
-
- Bembenek J, Yu H (2001) Regulation of the anaphase-promoting complex by the dual specificity phosphatase human Cdc14a. J Biol Chem 276: 48237–48242 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

