In vivo assembly and disassembly of Rad51 and Rad52 complexes during double-strand break repair
- PMID: 14765116
- PMCID: PMC380999
- DOI: 10.1038/sj.emboj.7600091
In vivo assembly and disassembly of Rad51 and Rad52 complexes during double-strand break repair
Abstract
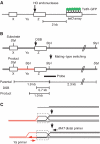
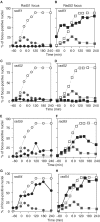
Assembly and disassembly of Rad51 and Rad52 complexes were monitored by immunofluorescence during homologous recombination initiated by an HO endonuclease-induced double-strand break (DSB) at the MAT locus. DSB-induced Rad51 and Rad52 foci colocalize with a TetR-GFP focus at tetO sequences adjacent to MAT. In strains in which HO cleaves three sites on chromosome III, we observe three distinct foci that colocalize with adjacent GFP chromosome marks. We compared the kinetics of focus formation with recombination intermediates and products when HO-cleaved MATalpha recombines with the donor, MATa. Rad51 assembly occurs 1 h after HO cleavage. Rad51 disassembly occurs at the same time that new DNA synthesis is initiated after single-stranded (ss) MAT DNA invades MATa. We present evidence for three distinct roles for Rad52 in recombination: a presynaptic role necessary for Rad51 assembly, a synaptic role with Rad51 filaments, and a postsynaptic role after Rad51 dissociates. Additional biochemical studies suggest the presence of an ssDNA complex containing both Rad51 and Rad52.
Figures






References
-
- Baumann P, West SC (1999) Heteroduplex formation by human Rad51 protein: effects of DNA end-structure, hRP-A and hRad52. J Mol Biol 291: 363–374 - PubMed
-
- Benson FE, Baumann P, West SC (1998) Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature 391: 401–404 - PubMed
-
- Bishop DK (1994) RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell 79: 1081–1092 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

