A Suv39h-dependent mechanism for silencing S-phase genes in differentiating but not in cycling cells
- PMID: 14765126
- PMCID: PMC1271807
- DOI: 10.1038/sj.emboj.7600074
A Suv39h-dependent mechanism for silencing S-phase genes in differentiating but not in cycling cells
Abstract
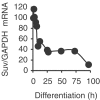
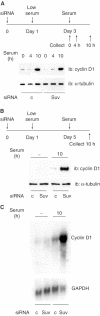
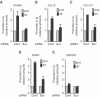
The Rb/E2F complex represses S-phase genes both in cycling cells and in cells that have permanently exited from the cell cycle and entered a terminal differentiation pathway. Here we show that S-phase gene repression, which involves histone-modifying enzymes, occurs through distinct mechanisms in these two situations. We used chromatin immunoprecipitation to show that methylation of histone H3 lysine 9 (H3K9) occurs at several Rb/E2F target promoters in differentiating cells but not in cycling cells. Furthermore, phenotypic knock-down experiments using siRNAs showed that the histone methyltransferase Suv39h is required for histone H3K9 methylation and subsequent repression of S-phase gene promoters in differentiating cells, but not in cycling cells. These results indicate that the E2F target gene permanent silencing mechanism that is triggered upon terminal differentiation is distinct from the transient repression mechanism in cycling cells. Finally, Suv39h-depleted myoblasts were unable to express early or late muscle differentiation markers. Thus, appropriately timed H3K9 methylation by Suv39h seems to be part of the control switch for exiting the cell cycle and entering differentiation.
Figures




References
-
- Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh PB, Reuter G, Jenuwein T (1999) Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J 18: 1923–1938 - PMC - PubMed
-
- Aagaard L, Schmid M, Warburton P, Jenuwein T (2000) Mitotic phosphorylation of SUV39H1, a novel component of active centromeres, coincides with transient accumulation at mammalian centromeres. J Cell Sci 113: 817–829 - PubMed
-
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124 - PubMed
-
- Brehm A, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T (1998) Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391: 597–601 - PubMed
-
- Buckingham M (1996) Skeletal muscle development and the role of the myogenic regulatory factors. Biochem Soc Trans 24: 506–509 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

