Chromate-reducing properties of soluble flavoproteins from Pseudomonas putida and Escherichia coli
- PMID: 14766567
- PMCID: PMC348923
- DOI: 10.1128/AEM.70.2.873-882.2004
Chromate-reducing properties of soluble flavoproteins from Pseudomonas putida and Escherichia coli
Abstract
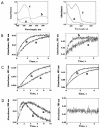
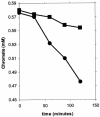
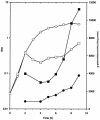
Cr(VI) (chromate) is a toxic, soluble environmental contaminant. Bacteria can reduce chromate to the insoluble and less toxic Cr(III), and thus chromate bioremediation is of interest. Genetic and protein engineering of suitable enzymes can improve bacterial bioremediation. Many bacterial enzymes catalyze one-electron reduction of chromate, generating Cr(V), which redox cycles, generating excessive reactive oxygen species (ROS). Such enzymes are not appropriate for bioremediation, as they harm the bacteria and their primary end product is not Cr(III). In this work, the chromate reductase activities of two electrophoretically pure soluble bacterial flavoproteins--ChrR (from Pseudomonas putida) and YieF (from Escherichia coli)-were examined. Both are dimers and reduce chromate efficiently to Cr(III) (kcat/Km = approximately 2 x 10(4) M(-1) x s(-1)). The ChrR dimer generated a flavin semiquinone during chromate reduction and transferred >25% of the NADH electrons to ROS. However, the semiquinone was formed transiently and ROS diminished with time. Thus, ChrR probably generates Cr(V), but only transiently. Studies with mutants showed that ChrR protects against chromate toxicity; this is possibly because it preempts chromate reduction by the cellular one-electron reducers, thereby minimizing ROS generation. ChrR is thus a suitable enzyme for further studies. During chromate reduction by YieF, no flavin semiquinone was generated and only 25% of the NADH electrons were transferred to ROS. The YieF dimer may therefore be an obligatory four-electron chromate reducer which in one step transfers three electrons to chromate and one to molecular oxygen. As a mutant lacking this enzyme could not be obtained, the role of YieF in chromate protection could not be directly explored. The results nevertheless suggest that YieF may be an even more suitable candidate for further studies than ChrR.
Figures

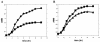

References
-
- Barceloux, D. G. 1999. Chromium. J. Toxicol. Clin. Toxicol. 37:173-194. - PubMed
-
- Cadenas, E. 1995. Antioxidant and prooxidant functions of DT-diaphorase in quinone metabolism. Biochem. Pharmacol. 49:127-140. - PubMed
-
- Campos-Garcia, J., G. Martinez-Cadena, R. Alvarez-Gonzalez, and C. Cervantes. 1997. Purification and partial characterization of a chromate reductase from Bacillus. Rev. Lat. Microbiol. 39:73-81. - PubMed
-
- Cervantes, C., J. Campos-Garcia, S. Devars, F. Gutierrez-Corona, H. Loza-Tavera, J. C. Torres-Guzman, and R. Moreno-Sanchez. 2001. Interactions of chromium with microorganisms and plants. FEMS Microbiol. Rev. 25:335-347. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

