Genetic diversity of Cryptosporidium spp. in captive reptiles
- PMID: 14766569
- PMCID: PMC348785
- DOI: 10.1128/AEM.70.2.891-899.2004
Genetic diversity of Cryptosporidium spp. in captive reptiles
Abstract
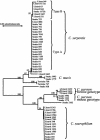
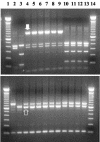
The genetic diversity of Cryptosporidium in reptiles was analyzed by PCR-restriction fragment length polymorphism and sequence analysis of the small subunit rRNA gene. A total of 123 samples were analyzed, of which 48 snake samples, 24 lizard samples, and 3 tortoise samples were positive for Cryptosporidium: Nine different types of Cryptosporidium were found, including Cryptosporidium serpentis, Cryptosporidium desert monitor genotype, Cryptosporidium muris, Cryptosporidium parvum bovine and mouse genotypes, one C. serpentis-like parasite in a lizard, two new Cryptosporidium spp. in snakes, and one new Cryptosporidium sp. in tortoises. C. serpentis and the desert monitor genotype were the most common parasites and were found in both snakes and lizards, whereas the C. muris and C. parvum parasites detected were probably the result of ingestion of infected rodents. Sequence and biologic characterizations indicated that the desert monitor genotype was Cryptosporidium saurophilum. Two host-adapted C. serpentis genotypes were found in snakes and lizards.
Figures


References
-
- Brownstein, D. G., J. D. Strandberg, R. J. Montali, M. Bush, and J. Fortner. 1977. Cryptosporidium in snakes with hypertrophic gastritis. Vet. Pathol. 14:606-617. - PubMed
-
- Fayer, R., U. Morgan, and S. J. Upton. 2000. Epidemiology of Cryptosporidium: transmission, detection and identification. Int. J. Parasitol. 30:1305-1322. - PubMed
-
- Fayer, R., C. Speer, and J. Dubey. 1997. The general biology of Cryptosporidium, p. 1-41. In R. Fayer (ed.), Cryptosporidium and cryptosporidiosis. CRC Press, Boca Raton, Fla.
-
- Graczyk, T. K., and M. R. Cranfield. 1998. Experimental transmission of Cryptosporidium oocyst isolates from mammals, birds and reptiles to captive snakes. Vet. Res. 29:187-195. - PubMed
-
- Graczyk, T. K., M. R. Cranfield, P. Helmer, R. Fayer, and E. F. Bostwick. 1998. Therapeutic efficacy of hyperimmune bovine colostrum treatment against clinical and subclinical Cryptosporidium serpentis infections in captive snakes. Vet. Parasitol. 74:123-132. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

