Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes
- PMID: 14766575
- PMCID: PMC348920
- DOI: 10.1128/AEM.70.2.943-953.2004
Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes
Abstract
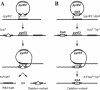
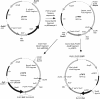
Since most archaea are extremophilic and difficult to cultivate, our current knowledge of their biology is confined largely to comparative genomics and biochemistry. Haloferax volcanii offers great promise as a model organism for archaeal genetics, but until now there has been a lack of a wide variety of selectable markers for this organism. We describe here isolation of H. volcanii leuB and trpA genes encoding 3-isopropylmalate dehydrogenase and tryptophan synthase, respectively, and development of these genes as a positive selection system. DeltaleuB and DeltatrpA mutants were constructed in a variety of genetic backgrounds and were shown to be auxotrophic for leucine and tryptophan, respectively. We constructed both integrative and replicative plasmids carrying the leuB or trpA gene under control of a constitutive promoter. The use of these selectable markers in deletion of the lhr gene of H. volcanii is described.
Figures




References
-
- Beggs, J. D. 1978. Transformation of yeast by a replicating hybrid plasmid. Nature 275:104-109. - PubMed
-
- Bell, S. D., and S. P. Jackson. 2001. Mechanism and regulation of transcription in archaea. Curr. Opin. Microbiol. 4:208-213. - PubMed
-
- Boeke, J. D., F. LaCroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345-346. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

