Biodegradation of the hexahydro-1,3,5-trinitro-1,3,5-triazine ring cleavage product 4-nitro-2,4-diazabutanal by Phanerochaete chrysosporium
- PMID: 14766596
- PMCID: PMC348896
- DOI: 10.1128/AEM.70.2.1123-1128.2004
Biodegradation of the hexahydro-1,3,5-trinitro-1,3,5-triazine ring cleavage product 4-nitro-2,4-diazabutanal by Phanerochaete chrysosporium
Abstract
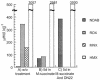
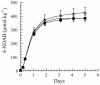
Initial denitration of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) by Rhodococcus sp. strain DN22 produces CO2 and the dead-end product 4-nitro-2,4-diazabutanal (NDAB), OHCNHCH2NHNO2, in high yield. Here we describe experiments to determine the biodegradability of NDAB in liquid culture and soils containing Phanerochaete chrysosporium. A soil sample taken from an ammunition plant contained RDX (342 micromol kg(-1)), HMX (octahydro-1,3,5,7-tetranitro-1,3,5,7-tetrazocine; 3,057 micromol kg(-1)), MNX (hexahydro-1-nitroso-3,5-dinitro-1,3,5-triazine; 155 micromol kg(-1)), and traces of NDAB (3.8 micromol kg(-1)). The detection of the last in real soil provided the first experimental evidence for the occurrence of natural attenuation that involved ring cleavage of RDX. When we incubated the soil with strain DN22, both RDX and MNX (but not HMX) degraded and produced NDAB (388 +/- 22 micromol kg(-1)) in 5 days. Subsequent incubation of the soil with the fungus led to the removal of NDAB, with the liberation of nitrous oxide (N2O). In cultures with the fungus alone NDAB degraded to give a stoichiometric amount of N2O. To determine C stoichiometry, we first generated [14C]NDAB in situ by incubating [14C]RDX with strain DN22, followed by incubation with the fungus. The production of 14CO2 increased from 30 (DN22 only) to 76% (fungus). Experiments with pure enzymes revealed that manganese-dependent peroxidase rather than lignin peroxidase was responsible for NDAB degradation. The detection of NDAB in contaminated soil and its effective mineralization by the fungus P. chrysosporium may constitute the basis for the development of bioremediation technologies.
Figures



References
-
- Ampleman, G., S. Thiboutot, J. Lavigne, A. Marois, J. Hawari, A. M. Jones, and D. Rho. 1995. Synthesis of 14C-labelled hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX), 2,4,6-trinitrotoluene (TNT), nitrocellulose (NC) and glycidylazide polymer (GAP) for use in assessing the biodegradation potential of these energetic compounds. J. Label. Compd. Radiopharm. 36:559-577.
-
- Balakrishnan, V., A. Halasz, and J. Hawari. 2003. Alkaline hydrolysis of the cyclic nitramine explosives RDX, HMX, and CL-20: new insights into degradation pathways obtained by the observation of novel intermediates. Environ. Sci. Technol. 37:1838-1843. - PubMed
-
- Bayman, P., S. D. Ritchey, and J. W. Bennett. 1995. Fungal interactions with the explosive RDX (hexahydro-1,3,5-trinitro-1,3,5-triazine). J. Ind. Microbiol. 15:418-423.
-
- Beller, H. R., and K. Tiemeier. 2002. Use of liquid chromatography/tandem mass spectrometry to detect distinctive indicators of in situ RDX transformation in contaminated groundwater. Environ. Sci. Technol. 36:2060-2066. - PubMed
-
- Bhushan, B., A. Halasz, J. Spain, S. Thiboutot, G. Ampleman, and J. Hawari. 2002. Biotransformation of hexahydro-1,3,5-trinitro-1,3,5-triazine catalyzed by a NAD(P)H: nitrate oxidoreductase from Aspergillus niger. Environ. Sci. Technol. 36:3104-3108. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

