Metamorphosis of a scleractinian coral in response to microbial biofilms
- PMID: 14766608
- PMCID: PMC348907
- DOI: 10.1128/AEM.70.2.1213-1221.2004
Metamorphosis of a scleractinian coral in response to microbial biofilms
Abstract
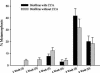
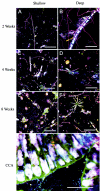
Microorganisms have been reported to induce settlement and metamorphosis in a wide range of marine invertebrate species. However, the primary cue reported for metamorphosis of coral larvae is calcareous coralline algae (CCA). Herein we report the community structure of developing coral reef biofilms and the potential role they play in triggering the metamorphosis of a scleractinian coral. Two-week-old biofilms induced metamorphosis in less than 10% of larvae, whereas metamorphosis increased significantly on older biofilms, with a maximum of 41% occurring on 8-week-old microbial films. There was a significant influence of depth in 4- and 8-week biofilms, with greater levels of metamorphosis occurring in response to shallow-water communities. Importantly, larvae were found to settle and metamorphose in response to microbial biofilms lacking CCA from both shallow and deep treatments, indicating that microorganisms not associated with CCA may play a significant role in coral metamorphosis. A polyphasic approach consisting of scanning electron microscopy, fluorescence in situ hybridization (FISH), and denaturing gradient gel electrophoresis (DGGE) revealed that coral reef biofilms were comprised of complex bacterial and microalgal communities which were distinct at each depth and time. Principal-component analysis of FISH data showed that the Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, and Cytophaga-Flavobacterium of Bacteroidetes had the largest influence on overall community composition. A low abundance of Archaea was detected in almost all biofilms, providing the first report of Archaea associated with coral reef biofilms. No differences in the relative densities of each subdivision of Proteobacteria were observed between slides that induced larval metamorphosis and those that did not. Comparative cluster analysis of bacterial DGGE patterns also revealed that there were clear age and depth distinctions in biofilm community structure; however, no difference was detected in banding profiles between biofilms which induced larval metamorphosis and those where no metamorphosis occurred. This investigation demonstrates that complex microbial communities can induce coral metamorphosis in the absence of CCA.
Figures

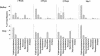


References
-
- Babcock, R. C., and A. J. Heyward. 1986. Larval development of certain gamete-spawning scleractinian corals. Coral Reefs 5:111-116.
-
- Brewer, R. H. 1976. Larval settling behaviour in Cyanea capillata (Cnidaria: Scyphozoa). Biol. Bull. 150:183-199.
-
- Characklis, W. G., and K. E. Cooksy. 1983. Biofilms and microbial fouling, p. 93-108. In A. I. Laskin (ed.), Applied microbiology. Academic Press, New York, N.Y.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

