High genetic diversity among Stenotrophomonas maltophilia strains despite their originating at a single hospital
- PMID: 14766838
- PMCID: PMC344440
- DOI: 10.1128/JCM.42.2.693-699.2003
High genetic diversity among Stenotrophomonas maltophilia strains despite their originating at a single hospital
Abstract
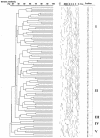
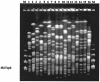
The levels of genetic relatedness of 139 Stenotrophomonas maltophilia strains recovered from 105 hospitalized non-cystic fibrosis patients (51% from medical wards, 35% from intensive care units, and 14% from surgical wards) and 7 environmental sources in the same hospital setting during a 4-year period were typed by the pulsed-field gel electrophoresis (PFGE) technique. A total of 99 well-defined distinct XbaI PFGE patterns were identified (Simpson's discrimination index, 0.996). The dendrogram showed a Dice similarity coefficient ranging from 28 to 80%. Two major clusters (I and II), three minor clusters (III, IV, and V), and two independent branches were observed when using a 36% Dice coefficient, indicating a high diversity of genetic relatedness. It is of note that 84% of strains were grouped within two major clonal lineages. No special cluster gathering was found among strains belonging to the same sample type specimen, patients' infection or colonization status, and ward of precedence. Despite this fact, three different clones (A, B, and C) recovered from respiratory samples from six, three, and two patients, respectively, and two clones, D and E, in two bacteremic patients each, were identified. Isolation of an S. maltophilia strain belonging to the clone A profile in a bronchoscope demonstrated a common source from this clone. This study revealed a high genetic diversity of S. maltophilia isolates despite their origin from a single hospital, which may be related to the wide environmental distribution of this pathogen. However, few clones could be transmitted among different patients, yielding outbreak situations.
Figures
References
-
- Arbeit, R. D., M. Arthur, R. Dunn, C. Kim, R. K. Selander, and R. Goldstein. 1990. Resolution of recent evolutionary divergence among Escherichia coli from related lineages: the application of pulsed field gel electrophoresis to molecular epidemiology. J. Infect. Dis. 161:230-235. - PubMed
-
- Betriu, C., A. Sánchez, M. L. Palau, M. Gómez, and J. J. Picazo. 2001. Antibiotic resistance surveillance of Stenotrophomonas maltophilia, 1993-1999. J. Antimicrob. Chemother. 48:152-154. - PubMed
-
- Cantón, R., S. Valdezate, A. Vindel, B. Sánchez del Salz, L. Máiz, and F. Baquero. 2003. Antimicrobial susceptibility profile of molecular typed cystic fibrosis Stenotrophomonas maltophilia isolates and differences with noncystic fibrosis isolates. Pediatr. Pulmonol. 35:99-107. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

