spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation
- PMID: 14766855
- PMCID: PMC344479
- DOI: 10.1128/JCM.42.2.792-799.2004
spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation
Abstract
Strain typing of microbial pathogens has two major aims: (i). to index genetic microvariation for use in outbreak investigations and (ii). to index genetic macrovariation for use in phylogenetic and population-based analyses. Until now, there has been no clear indication that one genetic marker can efficiently be used for both purposes. Previously, we had shown that DNA sequence analysis of the protein A gene variable repeat region (spa typing) provides a rapid and accurate method to discriminate Staphylococcus aureus outbreak isolates from those deemed epidemiologically unrelated. Here, using the hypothesis that the genetic macrovariation within a low-level recombinogenic species would accurately be characterized by a single-locus marker, we tested whether spa typing could congruently index the extensive genetic variation detected by a whole-genome DNA microarray in a collection of 36 isolates, which was recovered from 10 countries on four continents over a period of four decades, that is representative of the breadth of diversity within S. aureus. Using spa and coa typing, pulsed-field gel electrophoresis (PFGE), and microarray and multilocus enzyme electrophoresis (MLEE) data in molecular epidemiologic and evolutionary analyses, we determined that S. aureus likely has a primarily clonal population structure and that spa typing can singly index genetic variation with 88% direct concordance with the microarray and can correctly assign isolates to phylogenetic lineages. spa typing performed better than MLEE, PFGE, and coa typing in discriminatory power and in the degree of agreement with the microarray at various phylogenetic depths. This study showed that genetic analysis of the repeat region of protein A comprehensively characterizes both micro- and macrovariation in the primarily clonal population structure of S. aureus.
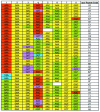
Figures

References
-
- Appel, R. D., A. Bairoch, and D. F. Hochstrasser. 1994. A new generation of information retrieval tools for biologists: the example of the ExPASy WWW server. Trends Biochem. Sci. 19:258-260. - PubMed
-
- Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. - PubMed
-
- Chang, S., D. M. Sievert, J. C. Hageman, M. L. Boulton, F. C. Tenover, F. P. Downes, S. Shah, J. T. Rudrik, G. R. Pupp, W. J. Brown, D. Cardo, and S. K. Fridkin. 2003. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N. Engl. J. Med. 348:1342-1347. - PubMed
-
- Crisostomo, M. I., H. Westh, A. Tomasz, M. Chung, D. C. Oliveira, and H. de Lencastre. 2001. The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clones. Proc. Natl. Acad. Sci. USA 98:9865-9870. - PMC - PubMed
-
- Day, N. P., C. E. Moore, M. C. Enright, A. P. Berendt, J. M. Smith, M. F. Murphy, S. J. Peacock, B. G. Spratt, and E. J. Feil. 2001. A link between virulence and ecological abundance in natural populations of Staphylococcus aureus. Science 292:114-116. (Retraction, 295:971, 2002.) - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

