The glyceryl ester of prostaglandin E2 mobilizes calcium and activates signal transduction in RAW264.7 cells
- PMID: 14766978
- PMCID: PMC357014
- DOI: 10.1073/pnas.0303950101
The glyceryl ester of prostaglandin E2 mobilizes calcium and activates signal transduction in RAW264.7 cells
Abstract
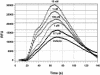
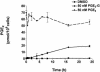
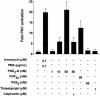
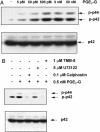
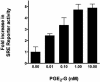
Glyceryl prostaglandins (PG-Gs) are generated by the oxygenation of the endocannabinoid, 2-arachidonylglycerol, by cyclooxygenase 2. The biological consequences of this selective oxygenation are uncertain because the cellular activities of PG-Gs have yet to be defined. We report that the glyceryl ester of PGE(2), PGE(2)-G, triggers rapid, concentration-dependent Ca(2+) accumulation in a murine macrophage-like cell line, RAW264.7. Ca(2+) mobilization is not observed after addition of PGE(2), PGD(2)-G, or PGF(2alpha)-G but is observed after addition of PGF(2alpha). Moreover, PGE(2)-G, but not PGE(2), stimulates a rapid but transient increase in the levels of inositol 1,4,5-trisphosphate (IP(3)) as well as the membrane association and activation of PKC. PGE(2)-G induces a concentration-dependent increase in the levels of phosphorylated extracellular signal regulated kinases 1 and 2 through a pathway that requires the activities of PKC, IP(3) receptor, and phospholipase C beta. The results indicate that PGE(2)-G triggers Ca(2+) mobilization, IP(3) synthesis, and activation of PKC in RAW264.7 macrophage cells at low concentrations. These responses are independent of the hydrolysis of PGE(2)-G to PGE(2).
Figures


References
-
- Smith, W. L. (1992) Am. J. Physiol. 263, F181–F191. - PubMed
-
- Yu, M., Ives, D. & Ramesha, C. S. (1997) J. Biol. Chem. 272, 21181–21186. - PubMed
-
- Kozak, K. R., Rowlinson, S. W. & Marnett, L. J. (2000) J. Biol. Chem. 275, 33744–33749. - PubMed
-
- Kozak, K. R., Crews, B. C., Morrow, J. D., Wang, L. H., Ma, Y. H., Weinander, R., Jakobsson, P. J. & Marnett, L. J. (2002) J. Biol. Chem. 277, 44877–44885. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

