The PXL1 gene of Saccharomyces cerevisiae encodes a paxillin-like protein functioning in polarized cell growth
- PMID: 14767053
- PMCID: PMC379286
- DOI: 10.1091/mbc.e04-01-0004
The PXL1 gene of Saccharomyces cerevisiae encodes a paxillin-like protein functioning in polarized cell growth
Abstract
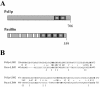
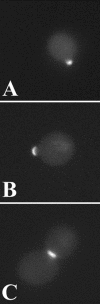
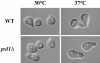
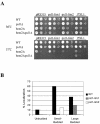
The Saccharomyces cerevisiae open reading frame YKR090w encodes a predicted protein displaying similarity in organization to paxillin, a scaffolding protein that organizes signaling and actin cytoskeletal regulating activities in many higher eucaryotic cell types. We found that YKR090w functions in a manner analogous to paxillin as a mediator of polarized cell growth; thus, we have named this gene PXL1 (Paxillin-like protein 1). Analyses of pxl1Delta strains show that PXL1 is required for the selection and maintenance of polarized growth sites during vegetative growth and mating. Genetic analyses of strains lacking both PXL1 and the Rho GAP BEM2 demonstrate that such cells display pronounced growth defects in response to different conditions causing Rho1 pathway activation. PXL1 also displays genetic interactions with the Rho1 effector FKS1. Pxl1p may therefore function as a modulator of Rho-GTPase signaling. A GFP::Pxl1 fusion protein localizes to sites of polarized cell growth. Experiments mapping the localization determinants of Pxl1p demonstrate the existence of localization mechanisms conserved between paxillin and Pxl1p and indicate an evolutionarily ancient and conserved role for LIM domain proteins in acting to modulate cell signaling and cytoskeletal organization during polarized growth.
Figures




References
-
- Alberts, A.S., Bouquin, N., Johnston, L.H., and Treisman, R. (1998). Analysis of RhoA-binding proteins reveals an interaction domain conserved in heterotrimeric G protein beta subunits and the yeast response regulator protein Skn7. J. Biol. Chem. 273, 8616-8622. - PubMed
-
- Altschul, S.F., Gish, W., Miller, W., Meyers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403-410. - PubMed
-
- Arkowitz, R.A. (1999). Responding to attraction: chemotaxis and chemotropism in Dictyostelium and yeast. Trends Cell Biol. 9, 20-27. - PubMed
-
- Ayscough, K.R. (2000). Endocytosis and the development of cell polarity in yeast require a dynamic F-actin cytoskeleton. Curr. Biol. 10, 1587-1590. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

