Identification of plastid envelope proteins required for import of protochlorophyllide oxidoreductase A into the chloroplast of barley
- PMID: 14769934
- PMCID: PMC380236
- DOI: 10.1073/pnas.0307284101
Identification of plastid envelope proteins required for import of protochlorophyllide oxidoreductase A into the chloroplast of barley
Abstract
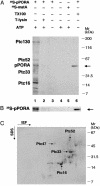
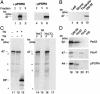
Chloroplasts synthesize an abundance of different tetrapyrrole compounds. Among them are chlorophyll and its precursor protochlorophyllide (Pchlide), which accumulate in light- and dark-grown plants, respectively. Pchlide is converted to chlorophyllide by virtue of the NADPH:Pchlide oxidoreductase (POR), which, in angiosperms, is the only known light-dependent enzyme of the chlorophyll biosynthetic pathway. In etiolated barley plants, two closely related POR proteins exist termed PORA and PORB, which are nuclear gene products. Here we identified plastid envelope proteins that interact with the cytosolic PORA precursor (pPORA) during its posttranslational chloroplast import. We demonstrate that pPORA interacts with several previously unreported components. Among them is a Pchlide a oxygenase, which provides Pchlide b as import substrate for pPORA, and a tyrosine aminotransferase thought to be involved in the synthesis of photoprotective vitamin E. Two other constituents were found to be orthologs of the GTP-binding proteins Toc33/34 and of the outer plastid envelope protein Oep16.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

