Inhibitors of protein synthesis identified by a high throughput multiplexed translation screen
- PMID: 14769948
- PMCID: PMC373382
- DOI: 10.1093/nar/gkh235
Inhibitors of protein synthesis identified by a high throughput multiplexed translation screen
Abstract
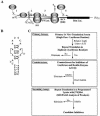
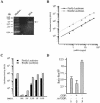
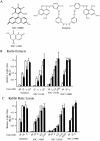
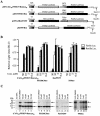
The use of small molecule inhibitors of cellular processes is a powerful approach to understanding gene function that complements the genetic approach. We have designed a high throughput screen to identify new inhibitors of eukaryotic protein synthesis. We used a bicistronic mRNA reporter to multiplex our assay and simultaneously screen for inhibitors of cap-dependent initiation, internal initiation and translation elongation/termination. Functional screening of >90 000 compounds in an in vitro translation reaction identified 36 inhibitors, 14 of which are known inhibitors of translation and 18 of which are nucleic acid-binding ligands. Our results indicate that intercalators constitute a large class of protein synthesis inhibitors. Four non-intercalating compounds were identified, three of which block elongation and one of which inhibits initiation. The novel inhibitor of initiation affects 5' end-mediated initiation, as well as translation initiated from picornaviral IRESs, but does not significantly affect internal initiation from the hepatitis C virus 5'-untranslated region. This compound should be useful for delineating differences in mechanism of initiation among IRESs.
Figures





References
-
- Pestka S. (1977) Inhibitors of Protein Synthesis. Academic Press, New York, NY.
-
- Vazquez D. (1979) Inhibitors of protein biosynthesis. Mol. Biol. Biochem. Biophys., 30, 1–312. - PubMed
-
- Graff J.R. and Zimmer,S.G. (2003) Translational control and metastatic progression: enhanced activity of the mRNA cap-binding protein eIF-4E selectively enhances translation of metastasis-related mRNAs. Clin. Exp. Metastasis, 20, 265–273. - PubMed
-
- Hershey J.W. and Miyamoto,S. (2000) Translation Control and Cancer. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

