In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer
- PMID: 14871930
- PMCID: PMC338285
- DOI: 10.1101/gad.1148404
In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer
Abstract
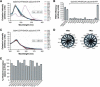
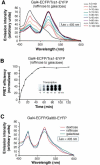
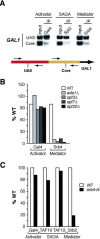
Our understanding of eukaryotic transcriptional activation mechanisms has been hampered by an inability to identify the direct in vivo targets of activator proteins, primarily because of lack of appropriate experimental methods. To circumvent this problem, we have developed a fluorescence resonance energy transfer (FRET) assay to monitor interactions with transcriptional activation domains in living cells. We use this method to show that the Tra1 subunit of the SAGA (Spt/Ada/Gcn5/acetyltransferase) complex is the direct in vivo target of the yeast activator Gal4. Chromatin-immunoprecipitation experiments demonstrate that the Gal4-Tra1 interaction is required for recruitment of SAGA to the upstream activating sequence (UAS), and SAGA, in turn, recruits the Mediator complex to the UAS. The UAS-bound Mediator is required for recruitment of the general transcription factors to the core promoter. Thus, our results identify the in vivo target of an activator and show how the activator-target interaction leads to transcriptional stimulation. The FRET assay we describe is a general method that can be used to identify the in vivo targets of other activators.
Figures




References
-
- Brown C.E., Lechner, T., Howe, L., and Workman, J.L. 2000. The many HATs of transcription coactivators. Trends Biochem. Sci. 25: 15-19. - PubMed
-
- Brown C.E., Howe, L., Sousa, K., Alley, S.C., Carrozza, M.J., Tan, S., and Workman, J.L. 2001. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292: 2333-2337. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
